Specimens of wild populations of the spotted rose snapper, Lutjanus guttatus (Steindacher) were studied for monogenean parasites in 2 localities along the Mexican Pacific coast (Mazatlán, Sinaloa and Chamela Bay, Jalisco). Five species of dactylogyrids were found on the gills of their hosts: Haliotrematoides guttati (García-Vargas, Fajer-Ávila, & Lamothe-Argumedo, 2008), H. plectridium Kristky and Mendoza-Franco in Kritsky, Tingbao, & Yuan, 2009, H. spinatus Kristky and Mendoza-Franco in Kritsky et al. (2009), Euryhaliotrema perezponceiGarcía-Vargas, Fajer-Ávila & Lamothe-Argumedo, 2008 and E. mehen (Soler-Jiménez, García-Gasca, & Fajer-Avila, 2012). Freshly collected specimens provided an opportunity to study and compare specimens from different localities in further detail and few morphological characters were added to the description of each species. Additionally, a fragment of 856bp of the 28S ribosomal RNA (D1–D3) was obtained for all the sampled monogeneans, and a phylogenetic analysis along with all available sequences of dactylogyrids was conducted to establish the systematic position of the species within the Ancyrocephalinae. Our results suggest that species of Haliotrema might be included in Haliotrematoides genus. In addition, the genetic divergence data suggest that H. guttati and H. spinatus may represent a species complex; however, this asseveration needs additional data.
Se estudiaron los monogéneos que parasitan poblaciones silvestres de “pargos” o “huachinangos”, Lutjanus guttatus (Steindacher) en 2 localidades de la costa del Pacífico mexicano (Mazatlán, Sinaloa y Bahía de Chamela, Jalisco). Se recolectaron 5 especies de dactylogíridos de las branquias de sus hospederos: Haliotrematoides guttati (García-Vargas et al., 2008), H. plectridium Kristky and Mendoza-Franco en Kristky, Tingbao, and Yuan (2009), H. spinatus Kristky and Mendoza-Franco en Kritsky et al. (2009), Euryhaliotrema perezponceiGarcía-Vargas, Fajer-Ávila & Lamothe-Argumedo, 2008 and E. mehen (Soler-Jiménez et al., 2012). El material permitió observar la morfología en mayor detalle y comparar ejemplares de diferentes localidades aportando algunos caracteres morfológicos a la descripción de las especies. Se obtuvo la secuencia de un fragmento de 856bp del gen ribosomal 28S (dominios D1 a D3), de este modo se estableció la posición sistemática de éstas, en conjunto con las secuencias de otros dactylogíridos, dentro de la filogenia de los Ancyrocephalinae. Este análisis reveló que las especies de Haliotrema podrían incluirse en el género Haliotrematoides. Asimismo, los datos de divergencia genética sugieren que H. guttati y H. spinatus podrían representar un complejo de especies, aunque esto requiere ser verificado con datos adicionales.
The genus Lutjanus (Steindacher) (Lutjanidae), the most species-rich among the family Lutjanidae, contains 68 species distributed around the world (Froese & Pauly, 2014). These fish are commercially important for fisheries, and at least 20 species have been examined for monogeneans, since it is well-known the impact that these parasites may have on fish populations. Species of 4 genera of dactylogyrids have been found on the gills of snappers around the world, EuryhaliotremaKritsky and Boeger, 2002, Haliotrema Johnston and Tiegs, 1922, HaliotrematoidesKritsky, Tingbao & Yuan, 2009 and Tetrancistrum Goto and Kikuchi, 1917; 2 other genera, Euryhaliotrematoides Plaisance and Kritsky, 2004, and Aliatrema Plaisance and Kritsky, 2004 were recently considered as synonyms of Euryhaliotrema based on the morphology of their male copulatory organ (MCO) (Kritsky, 2012). In Mexico, 9 dactylogyrid species have been described from snappers, i.e. H. cornigerum (Zhukov, 1976), H. gracilihamus (Zhukov, 1976), H. heteracantha (Zhukov, 1976), H. longihamus (Zhukov, 1976), H. magnigastrohamus (Zhukov, 1976), H. overstreeti Kritsky and Bullard in Kritsky et al. (2009) from Campeche Bay; H. guttati (García-Vargas, Fajer-Ávila & Lamothe-Argumedo, 2008), E. perezponceiGarcía-Vargas, Fajer-Ávila & Lamothe-Argumedo, 2008 from Mazatlán, Sinaloa and Cruz de Huanacaxtle, Nayarit, and E. mehen (Soler-Jiménez, García-Gasca & Fajer-Ávila, 2012) from Mazatlán, Sinaloa. The taxonomic status of some of these species of dactylogyrids has been unstable due to the lack of morphological details (mostly in haptoral hard parts and copulatory organ) in the original descriptions, although clarification has been gained after further taxonomic work conducted by several authors (e.g., Kritsky et al., 2009).
Recently, specimens of the spotted rose snapper Lutjanus guttatus (Steindachner) were collected in Chamela Bay, Jalisco and Mazatlán, Sinaloa, both on the Pacific coast of Mexico, as part of the study of the ectoparasites of fishes with commercial importance. From those fishes, specimens of dactylogyrids were collected from the gills. The objectives of this paper were to report the presence of 3 species of Haliotrematoides parasitizing the gills of spotted rose snappers, and to provide additional morphological information for each of them. In addition, we analyze the systematic position of these species (including E. perezponcei and E. mehen from Mazatlán) within the phylogeny of the Ancyrocephalinae, by using sequences of the 28S rRNA in the context of a wider phylogenetic analysis of dactylogyrids.
Materials and methodsSpecimens of adult spotted rose snapper Lutjanus guttatus were sampled in 2010 in Mazatlán Bay, Sinaloa (23°18′44″N, 106°29′37″W) and from 2011 through 2012 in Chamela Bay, Jalisco (19°33′15″N, 105°06′45″W), in northwestern Mexico; fish were obtained from local fishermen who set nets in each locality, kept on ice once removed from the nets, and examined for gill parasites a few hours after capture. Gills were extracted, fixed and stored in 96% ethanol; parasites were excised from the host tissue using triangular, surgical, mounted needles (size 16, Barber of Sheffield, U.K.) and were prepared for morphological and molecular evaluation. For morphological comparison, type-specimens deposited at the National Museum of Natural History (NMNH), Smithsonian Institution, Washington, D.C. (formerly U.S. National Parasite Collection – USNPC), USA at the Colección Nacional de Helmintos (CNHE) and at the Colección de Parásitos de Peces del Noroeste de México (CPPNP) were studied as follows: H. guttati (USNPC 101370–101371; CNHE, Neotype 8460), H. plectridium (USNPC 101367–101369), H. spinatus (USNPC 101353–101355; CNHE 6464–6465).
Morphological analysisThe haptor of each specimen was removed using a scalpel and air-dried on a glass slide; the corresponding body was transferred to a labeled Eppendorf tube containing 96% ethanol and stored at −20°C until required for molecular evaluation. Air-dried haptors were then subjected to a partial digestion using a proteinase K-base method (García-Vásquez, Hansen, Cristison, Bron, & Shinn, 2011). Some specimens were mounted (unstained) in Gray and Wess (Vidal-Martínez, Aguirre-Macedo, Scholz, González-Solis, & Mendoza-Franco, 2001) solution to clear the body tissue and visualize the haptoral hooks and copulatory complex. The haptoral armature and copulatory complex were studied using an immersion oil objective on an Olympus BX40 compound microscope. For comparisons, specimens were measured following Kritsky et al. (2009). When provided, measurements are presented in micrometers with the average followed by the range in parentheses. Specimens were deposited in the Colección Nacional de Helmintos from Mexico (CNHE) and in the U.S. National Parasite Collection (USNPC).
DNA extraction and sequencingParasite bodies were digested overnight at 56°C in a solution containing 10mM Tris–HCl (pH 7.6), 20mM NaCl, 100mM Na2 EDTA (pH 8.0), 1% Sakozyl, and 0.1mg/ml proteinase K. Following digestion, DNA was extracted from the supernatant using DNAzol reagent (Molecular Research Center, Cincinnati, Ohio) according to the manufacturer's instructions. Some tissues were extracted using the DNeasy Tissue Kit (Qiagen, Valencia, California). A fragment of the 28S ribosomal gene was amplified using the polymerase chain reaction (PCR). The primers Ancy55F (5′-GAGATTAGCCCATCACCG AAG-3′) (Plaisance, Littlewood, Olson, & Morand, 2005) and LSU1200R (5′-GCATAGTTCACCATCTTTCGG-3′) (Littlewood, Curini-Galletti, & Herniou, 2000) were used to amplify (PCR) a LSU fragment; also, the internal primers Halio-F (5′-ACCCGCTGAATTTAAGCAT-3′) and Halio-R (5′-TGGTCCGTGTTTCAAGAC-3′) (Soler-Jiménez et al., 2012). The PCR (25μl) consisted of 2.5μl of 1μl of 10μM of each primer, 2.5μl of 10× buffer, 1.5μl of MgCl2 15mM, 0.5μl dNTP's 10mM, 1U of Taq DNA polymerase (Platinum Taq, Invitrogen Corporation, São Paulo, Brazil), 2μl of DNA template, and 13.675μl of water. The PCR cycling protocol included denaturation at 94°C for 2min, followed by 35 cycles of 94°C for 1min, annealing at 50–58°C depending on dactylogyrid species (E. perezponcei 50°C, E. mehen 55°C, H. guttati and H. spinatus 58°C, and H. plectridium 55°C) for 1min and final extension at 72°C for 1min.
The reaction was performed in a MJ Research thermal cycler using a heated lid to reduce refluxing. Sequencing reactions were performed using an ABI Big Dye (Applied Biosystems, Boston, MA) terminator sequencing chemistry, and reaction products were separated and detected using an ABI 310 capillary DNA sequencer. Contigs were assembled and base calling differences resolved using Codoncode Aligner version 3.0.1 (Codoncode Corporation, Dedham, MA). All sequences were deposited in GenBank database.
Alignments and phylogenentic analysesSequences obtained in the current study from 28S (D1–D3) were aligned along with other species of Dactylogyridae available in GenBank using Clustal W implemented in Mega 5 (Tamura, Peterson, Peterson, Stecher, & Kumar, 2011) and adjusted manually with the Mesquite 2.75 program (Maddison & Maddison, 2011). Maximum likelihood (ML) and Bayesian Inference (BI) analyses were performed (Huelsenbeck & Ronquist, 2001). The jModelTest software version 0.1.1 (Posada, 2008) was used to select the best model of evolution for our dataset. The model (GTR+I+G) was selected based on the Akaike information criteria. The ML tree was inferred using RAxML 7.0.4, for each dataset (Stamatakis, 2006). Maximum likelihood clade support was assessed by bootstrap resampling with 10,000 replicates. Additionally, Bayesian analyses were performed with the program MrBAYES version 3.1.2 (Huelsenbeck & Ronquist, 2001). The settings were 2 simultaneous runs with 4 Markov chains and 5 million MCMC generations, sampling every 200 generations, a heating parameter value of 0.2 and a ‘burnin’ of 10%. Numbers at the interior branches of the consensus tree represent posterior probabilities (PP). Trees were drawn using FigTree program version 1.3.1 (Rambaut, 2006). The genetic divergence among species (E. perezponcei, E. mehen, H. plectridium, H. guttati and H. spinatus) was estimated using uncorrected “p-distances” method with the program MEGA v. 5 (Tamura et al., 2011).
ResultsThree species of Haliotrematoides were found parasitizing the gills of L. guttatus in Mazatlán, Sinaloa and Chamela Bay, Jalisco (Table 1). The specimens collected in the present study were compared morphologically with type and voucher specimens, mainly in terms of haptoral hooks and male copulatory organ (MCO). The following species were found:
Dactylogyrid species reported in the present study from the gills of Lutjanus guttatus from Chamela Bay, Jalisco and Mazatlán, Sinaloa, Mexico. CNHE-Coleccion Nacion de Helmintos, UNAM, México. USNPC – United States National Parasite Collection, USA.
| Monogenean | Locality | CNHE/USNPC | GenBank |
|---|---|---|---|
| Haliotrematoides guttati | Chamela Bay | 8354/106930 | KC663673–663674 |
| Mazatlán | 7293/– | HQ615993 | |
| Haliotrematoides plectridium | Chamela Bay | 8353/106931-106933 | – |
| Mazatlán | 7292/– | HQ615994–KC713622 | |
| Haliotrematoides spinatus | Chamela Bay | 8355/106934-106935 | KC663675–KC663678 |
| Mazatlán | 7291/– | HQ615995 |
Dactylogyridae Bychowsky, 1933
HaliotrematoidesKritsky et al., 2009
Haliotrematoides guttati (García-Vargas et al., 2008)
Taxonomic summarySite: gills.
Type host and locality: Lutjanus guttatus (Steindachner), Pacific Coast off Mazatlán, Sinaloa, Mexico (23°29′N, 106°36′W).
Host and localities: Chamela Bay, Jalisco, Mexico (19°31′49″N, 105°04′55″W). Mazatlán, Sinaloa (23°13′10″N, 106°26′05″W).
Previous records: Cruz de Huanacaxtle, Nayarit, Mexico (20°44′N, 105°22′W); Taboga Island, Panama (8°49′N, 79°34′W); Perlas Archipielago, Panama (8°22′N, 79°01′W).
Specimens deposited: CPPNP 7293, 5 vouchers (from the type locality), 4 vouchers CNHE 8354 and 1 voucher USNPC 106930 (from Chamela).
DNA reference sequence: the 856bp ribosomal DNA sequence of the LSU (D1-D3) is deposited in GenBank under accession numbers HQ615993 (from Mazatlán) and KC663673–KC663674 (from Chamela Bay).
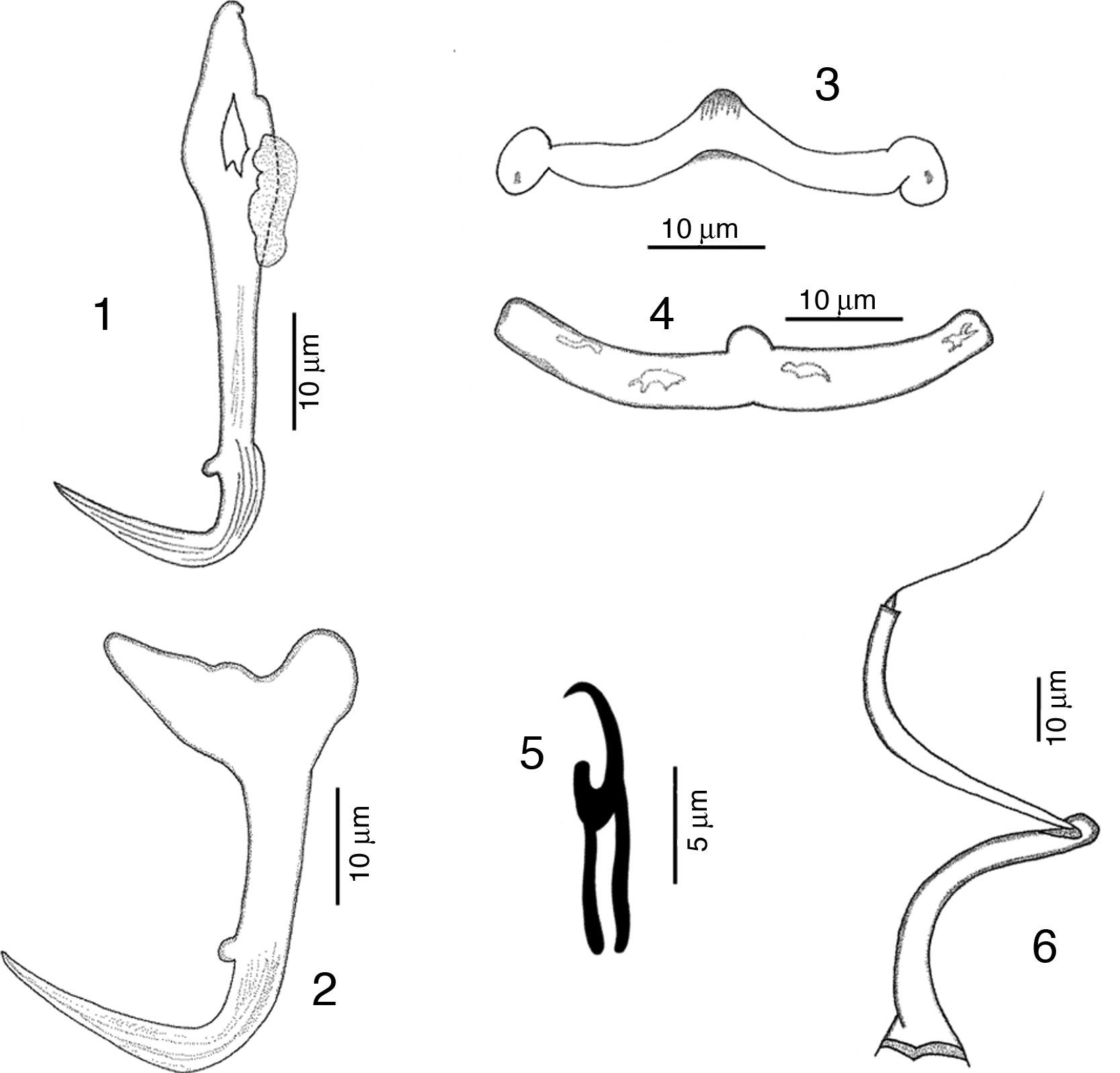
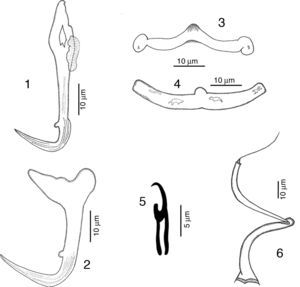
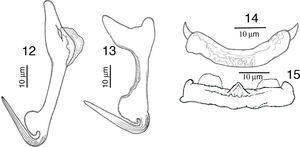
RemarksFive specimens of this species were found in Chamela bay, Jalisco and 10 were found in Mazatlán, Sinaloa, Mexico, corresponding with the genus description provided by Kritsky and Mendoza-Franco (Kritsky et al., 2009). This species was originally described as H. guttati as a parasite of L. guttatus from Mazatlán by García-Vargas et al. (2008). In 2009, Kritsky et al. found 2 specimens from the gills of snappers in Panama, and based on the comparison of their specimens with the original description, and their examination of the paratype and 2 voucher specimens (Harold W. Manter Laboratory, University of Nebraska-Lincoln, HWML, 48570, 48571) transferred the species to the genus Haliotrematoides. Following Kritsky et al. (2009) recommendation for the need of a re-description of this species, we studied in further detail our specimens and those deposited in museum collections (neotype, CNHE, and vouchers from the USNPC from Panama), and herein we provide the following details of some morphological traits: dorsal anchor with a well-developed membrane 12 (11–15) long, showing some striations on the surface of the curvature to the point; in the mid portion of perforation on the anchors base, a small heel is observed (not observed in specimens from Panama), a long inner root, no developed outer root (Fig. 1). Ventral anchor with both roots well developed, inner root 14 (12–16.5) long and ending pointed, longer than the outer root which is 6.5 (6–8) long and rounded (Fig. 2). Dorsal bar delicate, round in the middle, with a small knob, square ends where anchor attaches (Fig. 3). V-shaped ventral bar 41 (38.5–44) wide, but in description made by García-Vargas et al. (2008) is described as “W-shaped”, this was not observed in the type specimens that were studied (Fig. 4). Hooks with terminal depressed thumb directed to point, point opened and beyond thumb (Fig. 5). MCO tubular-shaped forming a spiral, conical base with the anterior part narrower than the base, and bearing a long whip heading to the anterior part of the body (Fig. 6).
Chamela Bay represents a new geographical record for this species.
Haliotrematoides plectridium Kritsky & Mendoza-Franco, 2009
Taxonomic summarySite: gills.
Type host and locality: Lutjanus guttatus (Steindachner), off Taboga Island, Panama (8°49′N, 79°34′W).
Host and localities: Lutjanus guttatus (Steinadachner), Chamela Bay, Jalisco, Mexico (19°31′49″N, 105°04′55″W), and Mazatlán, Sinaloa, Mexico (23°13′10″N, 106°26′05″W).
Previous records: Perlas Archipielago, Panama (8°22′N, 79°01′W).
Specimens deposited: CNHE 7292, 2 vouchers (from Mazatlán); CNHE 8353, 4 vouchers (from Chamela Bay).
DNA reference sequence: the 856bp ribosomal DNA sequence of the LSU (D1–D3) is deposited in GenBank under accession number HQ615994, KC713622 (from Mazatlán). No sequences were obtained from Chamela Bay, Mexico (10 specimens were prepared for molecular analysis but fail to amplify).
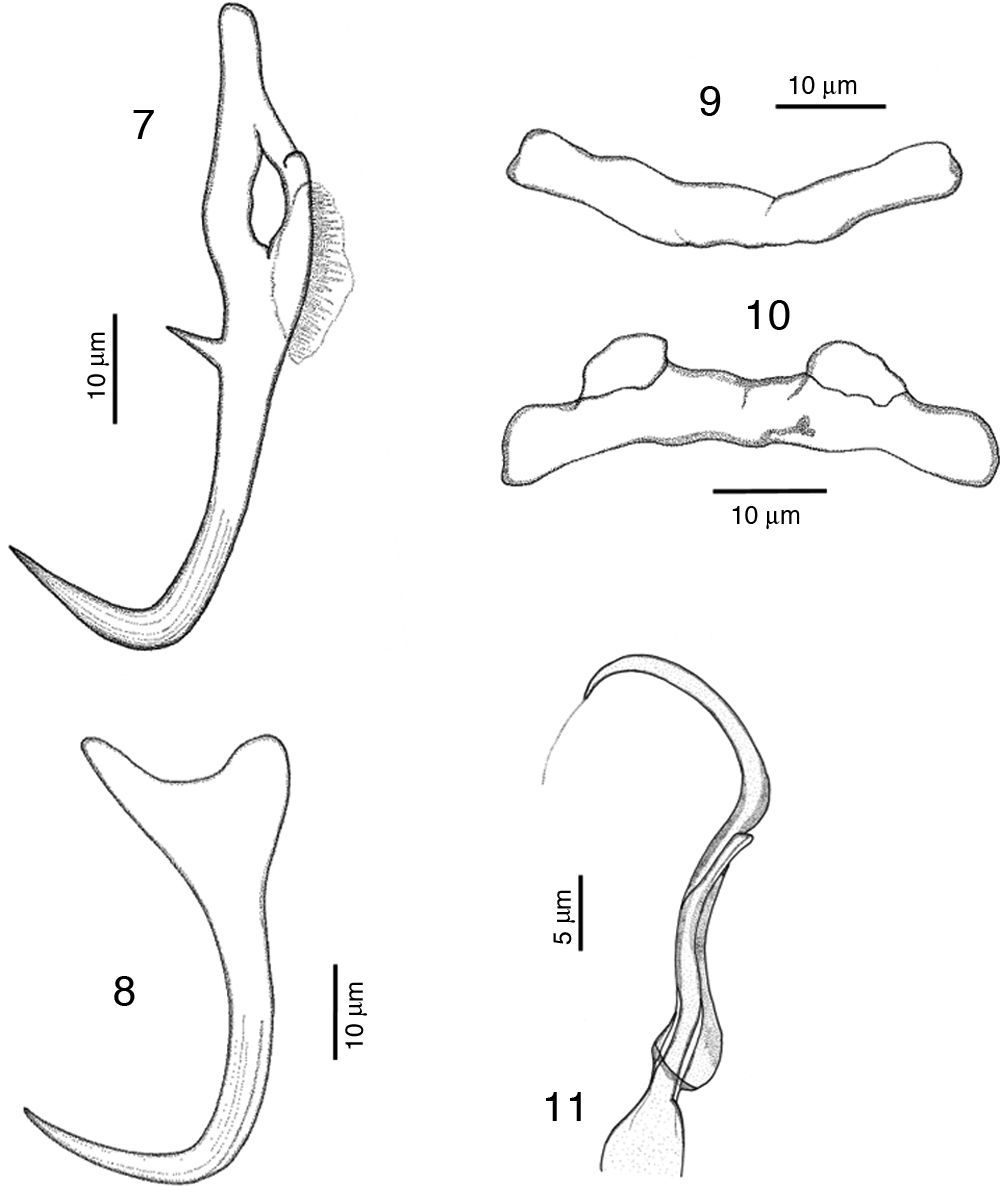
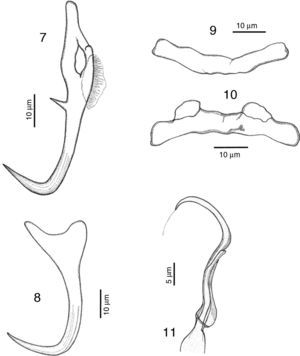
RemarksA total of 10 specimens were found in Mazatlán, Sinaloa and 19 in Chamela Bay, Jalisco. Our specimens are in agreement with the original description of Kritsky and Mendoza-Franco (2009) in Kritsky et al. (2009) from Taboga Island, and from Perlas Archipelago, both in Panama. Records from Mazatlán and Chamela represent new geographical records for this species. We observed in our specimens the following taxonomic traits, some of them slightly different from specimens from Panama: a membrane-like structure in the inner root of dorsal anchor, 8 (6–12.5) long, no outer root developed (Fig. 7). Ventral anchor with a depressed inner root, with the inner root longer than the outer root, 11 (8–17) long (Fig. 8). Dorsal bar 41 (36.5–46) wide, thin and concave while in the Panama specimens the bar is smaller, 35 (31–37) wide (Fig. 9). Ventral bar concave, 43 (40–47) wide, with 2 subterminal pockets in the anterior margin (convex in specimens from Panama) (Fig. 10). The MCO possess a filament at the end, finishing bulged in our specimens (Fig. 11), while this structure was not observed in Panama specimens.
Haliotrematoides spinatus Kritsky & Mendoza-Franco, 2009
Taxonomic summarySite: gills.
Type host and locality: Lutjanus guttatus (Steindachner), off Taboga Island, Panama (8°49′N, 79°34′W)
Host and localities: Chamela Bay, Jalisco, Mexico (19°31′49”N, 105°04′55”W), and Mazatlán, Sinaloa, México (23°29′N, 106°36′W).
Previous records: Perlas Archipielago, Panama (8°22′N, 79°01′W).
Specimens deposited: CNHE 7291 and CNHE 7546, 4 vouchers (from Mazatlán) and CNHE 8355, 4 vouchers, 2 vouchers USNPC 106934–106935 (from Chamela Bay).
DNA reference sequence: the 856bp ribosomal DNA sequence of the LSU (D1–D3) is deposited in GenBank under accession numbers HQ615995, KC713623–KC713627 (from Mazatlán) and KC663675–KC663678 (from Chamela Bay).
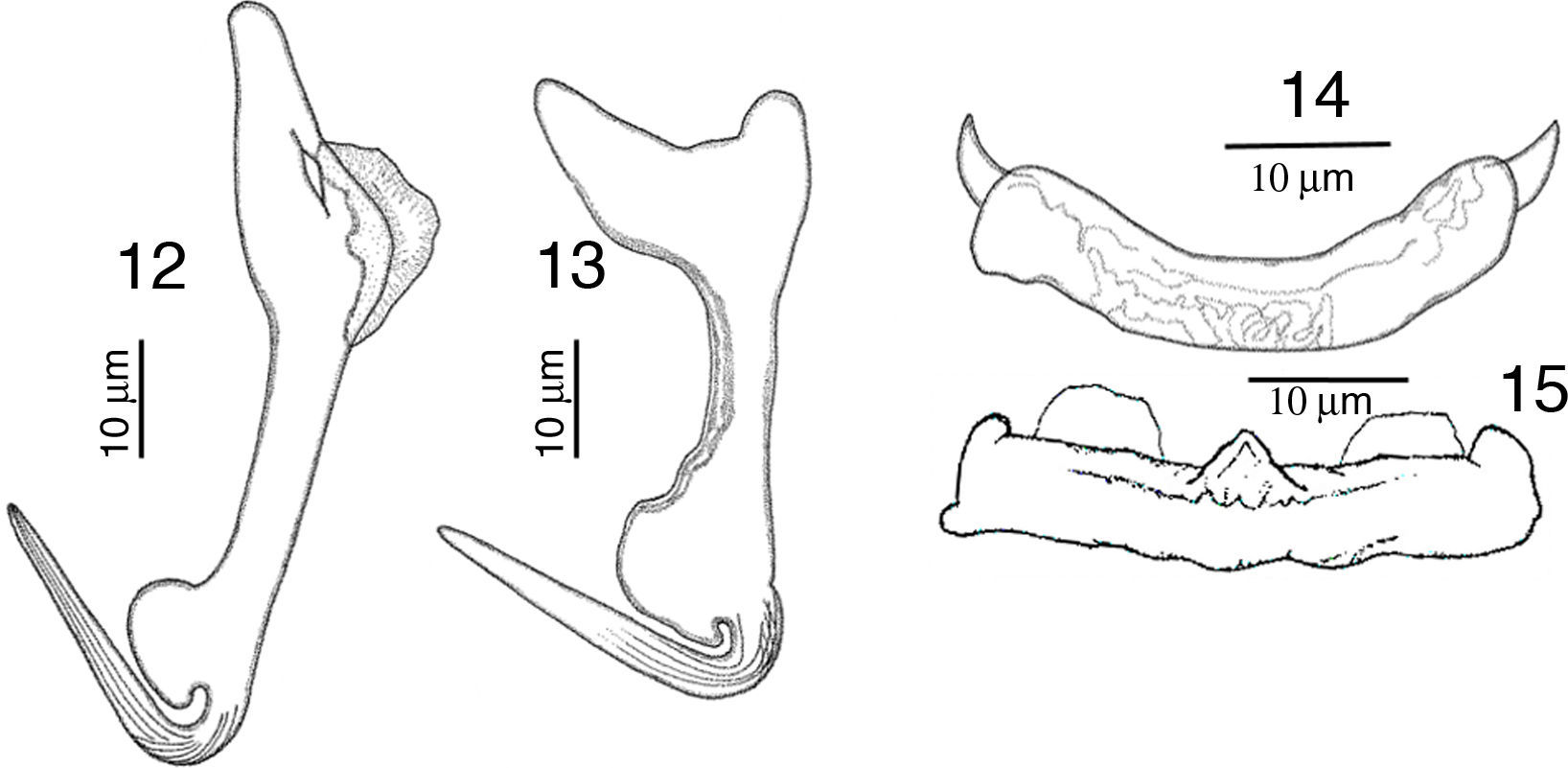
RemarksA total of 10 worms of this species were found in Mazatlán, Sinaloa and 11 from Chamela Bay, Jalisco. The specimens collected in the present study, conform to the description provided by Kritsky and Mendoza-Franco (2009) in Kritsky et al. (2009). The finding of this species in L. guttatus from Mazatlán and Chamela represent new geographical records. Our specimens show a delicate and short membrane in the outer root of the dorsal anchor, possessing a straight and relatively long point 24 (23–26) long, and a straight shaft (Fig. 12), meanwhile in the specimens from Panama the point is 21 long, and shaft is slightly curved, but differences might be due to intraspecific morphological variation. Ventral anchor with inner root small in our specimens, while in those from Panama the inner root is larger (Fig. 13). Dorsal bar slightly curved with short spine-like projections, anteriorly directed on each end, while specimens from Panama have 2 angled base dorsal bar (Fig. 14). The specimens of the present study possess a straight ventral bar, with 2 submedial pockets along the anterior margin and subterminal anterior notches (dorsal view), with a triangular ridge at the mid-portion of the anterior margin (Fig. 15), however in specimens from Panama this was not apparent.
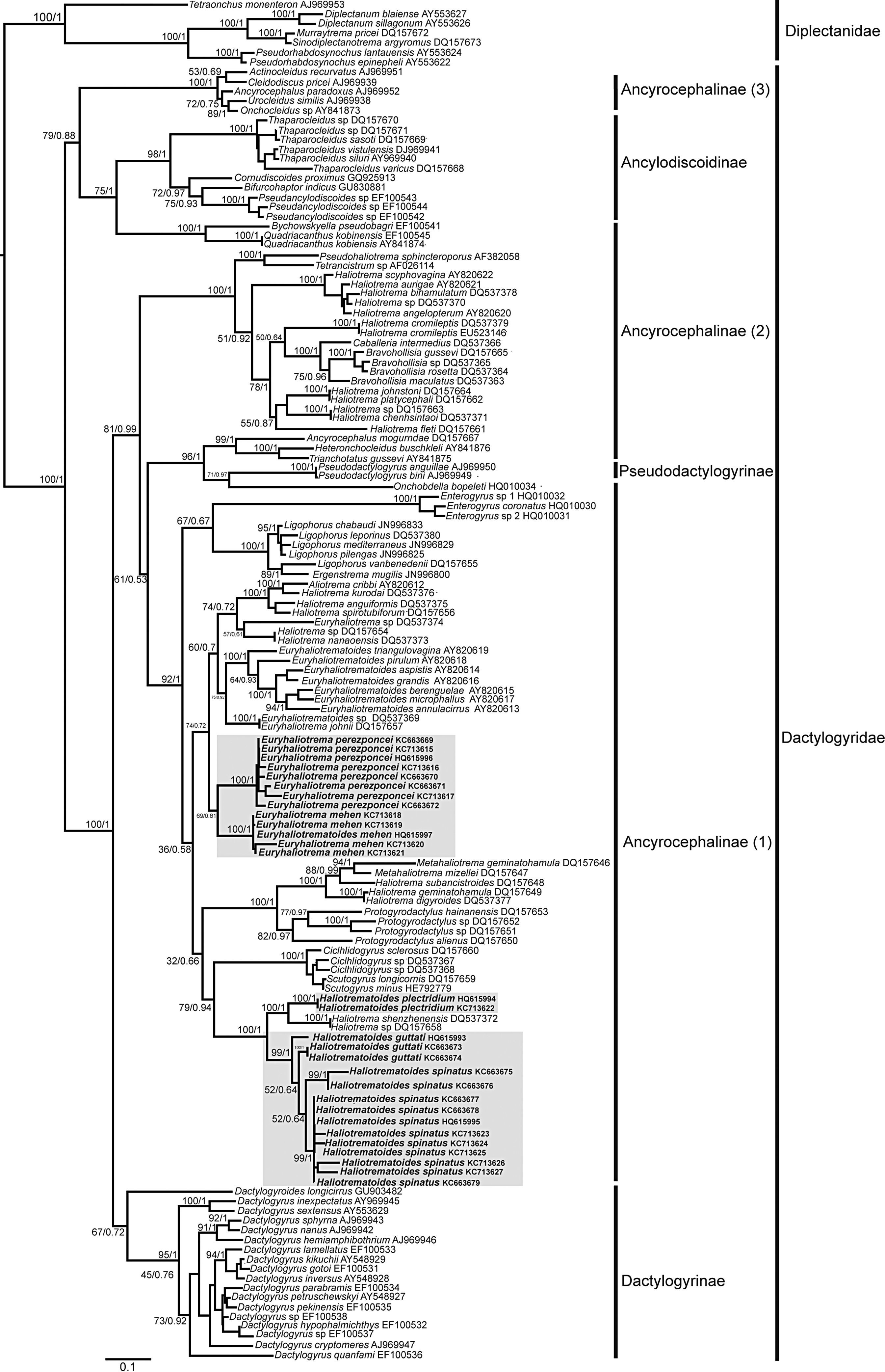
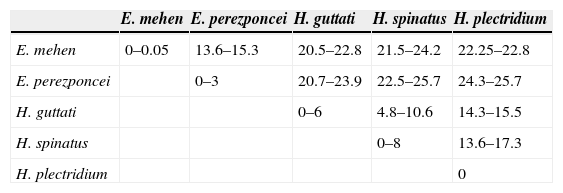
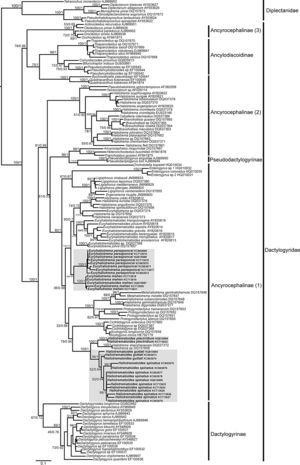
Systematic position and genetic divergence of Haliotrematoides spp.A total of 16 sequences were generated for the 3 species of Haliotrematoides recorded in this study: H. guttati (3 specimens), H. plectridium (2 specimens) and H. spinatus (11 specimens); additionally, 13 specimens of other 2 species of dactylogyrids were also sequenced: E. mehen (5), and E. perezponcei (8). The phylogenetic tree inferred from maximum likelihood analysis is shown in Fig. 16. The maximum likelihood tree (−ln likelihood=32,275.20) is presented, as the topology obtained from ML analysis was similar to the consensus tree of the Bayesian analysis. Likewise, the values of posterior probabilities are presented in the ML tree for comparison. The intra and interspecific genetic divergence (uncorrected P-distances) of species collected from snappers in Chamela Bay and Mazatlán is shown in Table 2. Euryhaliotrema and Haliotrematoides are not sister groups, and genetic divergence between genera varied from 20.5 to 24.2. The 3 species of Haliotrematoides differ in a range that varied from 4.8% between H. guttati and H. spinatus, and 17.3% between H. plectridium and H. spinatus. Intraspecific variation is very low, with the exception of that found among isolates of H. guttati (6%) and H. spinatus (8%). This intraspecific variation and the tree topology may indicate that these 2 species could represent a species complex. In the first case, the 3 isolates of H. guttati are not monophyletic, although this relationship is supported by low bootstrap and posterior probability values. Instead, the isolates of H. spinatus conform a monophyletic clade, although 2 subclades are formed, each with high support values (99/1) (Fig. 16). Even though this could represent a case for the existence of cryptic species, since morphological characters distinguishing those groups are not observed, the inclusion of other molecular markers and a larger sample size is necessary to test the hypothesis of a species complex.
Uncorrected pairwise distances of the partial sequences of the 28S rRNA gene among species analyzed from Chamela Bay, Jalisco and Mazatlán, Sinaloa, Mexico. Values are given as a percentage (%).
| E. mehen | E. perezponcei | H. guttati | H. spinatus | H. plectridium | |
|---|---|---|---|---|---|
| E. mehen | 0–0.05 | 13.6–15.3 | 20.5–22.8 | 21.5–24.2 | 22.25–22.8 |
| E. perezponcei | 0–3 | 20.7–23.9 | 22.5–25.7 | 24.3–25.7 | |
| H. guttati | 0–6 | 4.8–10.6 | 14.3–15.5 | ||
| H. spinatus | 0–8 | 13.6–17.3 | |||
| H. plectridium | 0 |
Species of the genus Haliotrematoides herein studied are nested within the Ancyrocephalinae sensu lato (Fig. 16). Sequences of 2 species of Haliotrema described as parasites of Sciaenops ocellatus L. (Sciaenidae) in China (one of them H. schenzhenensis, Wang, Lui, & Zhou, 2003), are nested as the sister group of H. plectridium, a relationship highly supported by bootstrap and posterior probability values. We were unable to confirm the taxonomic determination of that species since we had no access to museum specimens. This is what the DNA sequence data suggest, however this need to be taken with caution, because it is possible that the species from China may actually belong to the genus Haliotrematoides, but this needs to be determined when type-specimens are studied.
DiscussionIn this study new geographical records of 3 species of Haliotrematoides are presented, and a few details of the morphology of these species are given to complete their taxonomic description. In addition, the specimens collected were sequenced and new data was generated for the 28S rRNA gene, establishing sister-group relationships of the genus within the Ancyrocephalinae sensu lato. Genetic divergence values indicate that the species H. guttati and H. spinatus potentially represent a species complex. Unfortunately, we found no characters that allowed us to distinguish the specimens on morphological grounds. However, since we used in this study only 1 molecular marker, we took a conservative position and we decided to only recognize the potential existence of 2 cryptic species. These results need to be further corroborated by using at least another molecular marker such as cox1, which is more variable and may be more precise in identifying genetic lineages corresponding with separate species, and also it has been proven to be useful in achieving comprehensive species descriptions that facilitate reliable species diagnostics and rapid assessment of biodiversity, as in the case of species in the genus Gyrodactylus von Nordmann, 1832 (Hansen, Bakke, & Bachmann, 2007).
A single molecular marker has been used in previous studies on monogenoids when characterizing new species or describing genetic divergence; some studies used either a ribosomal marker (such as internal transcribed spacers) (Huyse & Volckaert, 2002), or a mitochondrial gene such as cox1 (Hansen, Bachmann, & Bakke, 2003). However, when the potential presence of cryptic species is recognized (Nadler & Pérez-Ponce de León, 2011; Pérez-Ponce de León & Nadler, 2010), the use of several molecular markers in the context of a phylogenetic analysis, and the proper characterization of genetic divergence is highly recommended. This approach has been followed in some species of monogenoids by authors such as Ziętara and Lumme (2003) and Kuusela, Ziętara, and Lumme (2008).
The partial fragment of the 28S rRNA (D1-D3) gene has been widely used to investigate the phylogenetic relationships among some dactylogyrids (Blasco-Costa, Miguez-Lozano, Sarabeev, & Balbuena, 2012; Dang, Levsen, Shander, & Bristow, 2010; Plaisance et al., 2005; Šimková, Mat¿jusová, & Cunningham, 2006; Singh & Chaudhary, 2010, 2011). Our results support the classification scheme proposed by Kritsky and Boeger (1989), who described the polyphyletic and paraphyletic nature of this group of monogenoids. In our study, Ancyrocephalinae was also not recovered as a monophyletic group.
As an additional result of our research, sequences of specimens of other ancyrocephalid monogeneans infecting the spotted rose snapper were obtained, particularly specimens of E. perezponcei that were collected from the gills of L. guttatus, a species previously described by García-Vargas et al. (2008), as well as specimens of E. mehen, a species described by Soler-Jiménez et al. (2012), both from Mazatlán. The taxonomic status of the genus Euryhaliotema has been also unstable. The genus was proposed by Kritsky and Boeger (2002) and, in a recent review of the genus by Kritsky (2012), the author discovered that some specimens collected from snappers possessed a mixture of morphological features, and decided to include the species of Euryhaliotrematoides within the genus Euryhaliotrema. The species described by Soler-Jiménez et al. (2012) as Euryhaliotrematoides mehen infecting L. guttatus from Mazatlán was then transferred to Euryhaliotrema by Kritsky (2012). The molecular results we obtained herein confirm the proposal by Kritsky (2012) since E. perezponcei nests as the sister species of E. mehen. The presence of E. perezponcei as a parasite of L. guttatus in Chamela represents a new geographical record, and the fact that sequences of individuals of this species from Mazatlán and Chamela Bay are nearly identical may indicate that, irrespective of the geographical distance between localities, populations of the spotted rose snapper move along the Pacific coast of Mexico.
We would like to thank the following people for the loan of specimens of H. guttati, H. plectridium and H. spinatus, Eric Hoberg and Patricia Pilitt from United States National Parasite Collection, Luis García Prieto from the CNHE and Rosy Medina from the CPPNP; to fishermen in Chamela Bay, Jalisco, Mazatlán and Sinaloa, for their help to obtain the fish. To Berenit Mendoza Garfias, Rosario Briosio Aguilar, Neptalí Morales Serna, Aline Rojas Sánchez and Rosy Medina for their help during field work. To Martín García Varela for his assistance with molecular analysis. This work was partly supported by PAPIIT IN204514 to GPPL. AGV thanks DGAPA-UNAM for a Postdoctoral scholarship.
Peer Review under the responsibility of Universidad Nacional Autónoma de México.