Periodontal ligament is a fibrous connective tissue of ectomesenchymal origin which contains a multipotent postnatal stem cells population with clonogenic capacity and high proliferation index. Multipotent postnatal stem cells population culture has been achieved through several extraction methods, nevertheless some technical problems have been encountered. For this reason, in the present study, 6 different methods were compared so as to improve isolation of primary periodontal ligament cells. Multipotent postnatal stem cells population were isolated from 36 healthy premolars, which were then cultured in complete Dulbecco modified Eagle medium (BioWest) (added with L-glutamine, 10% fetal bovine serum and 1% antibiotic), in standard culture conditions. Assays for cell proliferation were conducted with 3-(4,5 dimethylthiazol-2-yl)-2,5 diphenyltetrazolium technique as well as cell migration through crop injury test, after which osteogenic differentiation was assessed. Cultures were performed in triplicate with each of the published techniques; thus we were able to obtain a minimum population of cells susceptible to contamination. The «simplified method» was conducted through explant enzymatic digestion and culture: with obtained multipotent postnatal stem cells population, a 14 day proliferation was observed in standard conditions of ascendant culture; in migration tests, multipotent postnatal stem cells population totally covered injured surfaces after 72hours. CD90 positive expression was observed in over 80% of all cells; CD73 positive expression was found in 70% of cells, CD105 in 60% and vimentin in 90% of the population, as well as negative marking to CD11b and CD45. Positive osteogenic differentiation was observed after 14 days, through positive expression to RUNX2 in at least 15% of cells, and to osteocalcin in 90% as well as positive test to alizarin red. Based on obtained results we were able to affirm that the simplified method allows to isolate a population of mesenchymal cells, which might be obtained from extracted healthy premolars.
El ligamento periodontal es un tejido conectivo fibroso, de origen ectomesenquimal que contiene una población de células troncales postnatales multipotenciales, con capacidad clonogénica y alto índice de proliferación. Se ha conseguido cultivar células troncales postnatales multipotenciales a través de diferentes métodos de extracción, con algunos problemas técnicos. Por esta razón, se compararon seis diferentes métodos a fin de mejorar el aislamiento de células primarias del ligamento periodontal. Se aislaron células troncales postnatales multipotenciales a partir de 36 premolares sanos, que se cultivaron en medio de Eagle modificado por Dulbecco (BioWest) completo (adicionado con L-glutamina, 10% de Suero fetal bovino y 1% de antibiótico) y en condiciones estándares de cultivo. Se realizaron los ensayos para proliferación celular mediante la técnica de 3-(4,5-dimetiltiazol-2-il)-2,5-difeniltetrazolio, migración celular a través de la prueba de lesión del cultivo, y se evaluó su diferenciación osteogénica. Los cultivos realizados por triplicado con cada una de las técnicas publicadas, nos condujo a obtener una población mínima de células que fueron susceptibles a contaminarse. Se realizó con el «método simplificado»: por digestión enzimática y cultivo de explantos, con las células troncales postnatales multipotenciales obtenidas se observó una proliferación durante 14 días en condiciones estándares de cultivo ascendente, en las pruebas de migración las células troncales postnatales multipotenciales cubrieron totalmente la superficie lesionada a las 72 horas, se observó expresión positiva de CD90 en más del 80% de las células; expresión positiva a CD73 en un 70%, CD105 en un 60% y vimentina en un 90% de la población, así como un marcaje negativo a CD11b y CD45. Se observó una diferenciación osteogénica positiva a los 14 días mediante la expresión positiva a RUNX2 en al menos el 15% de las células y a osteocalcina en un 90%, así como las pruebas positivas a rojo de alizarina. Con los resultados obtenidos, podemos afirmar que el método simplificado permite aislar una población de células mesenquimales, que se pueden obtener a partir de premolares sanos extraídos.
Acronyms: ALP = Alcaline phosphatase, DMEM = Dulbecco modified Eagle medium, FBS = Fetal bovine serum, MSC = Mesenchymal stem cells, MTT = 3-(4,5 dimethylthiazol-2-il)-2,5 diphenyltetrazolium, PBS = Phosphate buffered saline solution, PDL = Periodontal ligament, PDLSC = Periodontal ligament stem cells, PLCSC = Peripheral limbar cornea stromal cells.
INTRODUCTIONPeriodontal ligament is a fibrous connective tissue composed of fibers, cell and collagen-rich intercellular substance, protein content and polysaccharides, it is of ecto-mesenchymal origin. It encompasses the following functions: 1) suspend the tooth in its bone socket, 2) provide nutrients to both socket and cement, 3) protect the teeth, and 4) preserve homeostasis of teeth within the socket by means of continuous regeneration.1 This regeneration of the Periodontal Ligament (PDL) as a continuous physiological mechanism, gave rise to the theory that this tissue could contain a subpopulation of stem cells.2 In 2004, Seo et al3 published that it was possible for human PDL to contain a population of multipotential, postnatal stem cells, which would possess clonogetic capacity, high proliferation index and the ability to regenerate periodontal tissue.4–6
It has been shown that mesenchymal periodontal ligament stem cells (PDLSCs) are similar to other mesenchymal stem cells in the expression of the following markers: STRO-1, CD146, CD90, CD29, CD44, CD13, CD105 and CD166, as well as their self-regenerating capacity and their multi-potentiality.3,7,8 Several authors, after isolating PDLSCs showed their ability to differentiate to mesodermal type lineages, differentiating in cementoblast-like cells, osteoblasts, chondrocytes, adipocytes, myofibroblasts, and collagen-forming fibers.9,10 Potential of PDLSCs has even been researched to give rise to nervous system-like cells and cementoblasts.11
Presently, PDLSCs haven been successfully cultured through different extraction methods, showing that PDL is a suitable source of stem cells, with the consequent potential use in regenerative medicine. Nevertheless, methods described to this date encounter some technical problems. For this reason, six methods were compared (Table I) in an effort to improve isolation methods of PDL primary cells, since none of the reported methods was precise. The aim of the present research paper was to present a highly replicable, simplified method for harvesting the aforementioned cells.
Comparative and explanatory table of techniques used for cell extraction (harvesting better?). Differences among them are underlined.
| Method | Authors | Transport and previous treatment | Procedure | Culture | Maintenance | Advantages | Disadvantages |
|---|---|---|---|---|---|---|---|
| Enzymatic digestion with type 1 collagenase | Vasandan AB, Shankar SR, Prasad P et al, 2013 (606 Vasandan AB, 2014) | In PBS at 1x added with 1% antibiotic/antifungal, 4 additional washes with this solution | 500μL type I collagenase at a concentration of 0.5mg/mL during 12hours at 37°C | Inactivated with 1mL complete D-MEM. Centrifuged at 1200 RPM during 10minutes, supernatant was decanted, sediment tissue was re-suspended with 2mL complete D-MEM | In 6 well plates, 300μL suspension in each well, under standard conditions, medium was changed 24hours after culture. Incubation culture medium was changed every 72hours | Conducted rinses are efficient | Excessive time of enzymatic digestion |
| Enzymatic digestion with type I collagenase | Seo BM, Miura M, Gronthos S et al, 2004 (607 Seo BM, 2004) | Complete DS-MEM plus 3washes during 3minutes | 500μL of type I collagenase at a 0.5mg/mL concentration during 1 hour at 37°C | Reaction was inactivated by adding 1mL of complete D-MEM, suspension was filtered with a 70μm diameter mesh, it was centrifuged at 1800 RPM during 15minutes, sediment was re-suspended with 2mL of complete D-MEM | 500μL of suspension were seeded in 6-well plates, under standard culture conditions. Culture medium was changed every 72hours | Sample vitality during transportation with D-MEM | Tissue loss during filtering, the first period to change culture medium is excessive |
| Mechanical disaggregation, explant culture | Tanaka K, Iwasaki K, Feghali KE at al, 2011 (608 Tanaka K, 2011) | Complete D-MEM, 10% FBS, 1% L-glutamine and 0.1% antibiotic/antifungal plus 5 washes during 30 seconds | Tissue fragments of approximately 1mm diameter were cut (explants) | Explants were strategically placed in 6 well boxes with a minimum amount of complete D-MEM L | Explants were incubated under standard conditions during a 6-8 hour period to wait for their adhesion to the culture plate. After this 500 μL of complete D-MEM were added. Medium was changed after 24hours to eliminate floating tissue, and after that at 72 hour intervals | Use of explants provided high expectancy to obtain primary tissue | Washes with deficient time. Very prolonged time for explant adhesion |
| Enzymatic digestion with trypsin/EDTA 0.25%/0.1% | Tanaka K, Iwasaki K, Feghali KE at al, 2011 (608 Tanaka K, 2011) | Complete D-MEM, 10% FBS, 1% L-glutamine and 0.1% of antibiotic/antifungal plus two washes during 2 minutes | Tissue was digested during 60minutes at 37°C in 500 μL of 0.25% trypsin with 0.1% EDTA. At 10 minute intervals the sample was shaken during 30 seconds | 500 μL complete D-MEM were added to arrest enzymatic digestion. Suspension was filtered with a 70 μm diameter mesh. Suspension was centrifuge at 1800 RPM during 15minutes. Supernatant was discarded and sediment was re-suspended with 2mL complete D-MEM | In six well plates, 500 μL of suspension were placed in each well. Medium was changed every 72 hours | With use of trypsin/EDTA enzymatic digestion was enhanced with constant shaking | Deficiencies in the washes. Prolonged time for enzymatic digestion, sample filtering |
| Enzymatic digestion with type I collagenase and sample transport in Hanks solution | Gholamrezanezhad Ali, 2011 (609, Gholamrezanezhad Ali, 2011) | Hanks solution at a 4°C temperature, three 1 minute washes | Tissue was centrifuged with Hanks solution at 1500 RPM for 10minutes. Supernatant was discarded and tissue was re-suspended with 500 μL type I collagenase at 0.5 mg/mL concentration for one hour at 37°C | Reaction inactivation was achieved with one milliliter of complete D-MEM. Suspension was centrifuged at 1500 RPM during 10minutes. Supernatant was discarded and sediment was re-suspended with 2mL of complete D-MEM | 200 μL of suspension were culture in 6 well plates during 4hours at 37°C. After this, 500 μL of complete D-MEM were added. Medium was substituted for the first time at 5 days, and after that every 72 hours | Adequate incubation time to wait for explant adhesion | Transport in Hanks solution. Deficient washes |
| Standardized technique enzymatic digestion and explant culture | Ramírez RO, Trejo ICG, 2015 | Complete D-MEM at a temperature of 4°C. Three 5 minute washes were performed with 3mL D-MEM supplemented with 5% antibiotic | Enzymatic digestion with 500 μL of trypsin/EDTA 0.25%/0.1% during 20minutes at 37°C. At five minute intervals, tissue was shaken with a Vortex at 1000 RPM during 30 seconds | Reaction inactivation was achieved with 500 μL of complete D-MEM. Centrifugation at 1800 RPM during 10minutes, supernatant was discarded and tissue was resuspended with 1mL complete D-MEM | 150-200 μL suspension were seeded in 6 wells, incubation was allowed for 2-4hours waiting for adhesion of tissue to the culture plate. After this, complete D-MEM was added and medium was changed 4 days after seeding, after this, medium was changed every 72 h | Advantages of previous techniques were enjoyed |
Thirty six healthy premolars were obtained; they were donated by patients of the Naucalpan Orthodontic Specialty Clinic, who had previously signed informed consent forms according to the Bioethics Committee of the Iztacala Graduate School, attached to the National Autonomous University of Mexico (UNAM). The following inclusion criteria were used for tooth selection: only patients without systemic alterations, suitable periodontal health, first and second premolar teeth from right and left upper and lower jaws. Exclusion criteria were: patients afflicted with systemic diseases, patients with gingivitis and/or periodontal disease, premolars with presence of dental plaque and/or presence of caries and/or periapical reaction.
Samples were transferred in 3mm of D-MEM medium (BioWest) supplemented with 1% antibiotic (penicillin-streptomycin Gibco® N° Cl. 15140-122) at 4°C.
Employed techniques are meticulously described in table I. In all seeding techniques complete D-MEM (BioWest) was used (added with L-glutamine, 10% of FBS and 1% of antibiotic, in standard culture conditions at 37°C with presence of 5% CO2.
Subcultures were performed with 60/80% confluence, after having been washed for 30seconds with Hank solution (Sigma Aldrich); 500μL of tripsin/ EDTA at 0.25%/0.1% (BioWest) during 1-3minutes at a 37°C temperature. Cell subcultures were conducted in a 1/3 proportion.
Cell proliferation assayFive hundred cells were seeded in 24-well boxes, one cell to a well. Assessment of cell proliferation was conducted at 1, 3, 5, 7, and 14 days after seeding, this was accomplished with addition of 50μL of MTT (Sigma Aldrich) incubated for four hours at 37°C in a 5% CO2 atmosphere. At a later point, 100μL of dimethyl sulfoxide (DMSO, Sigma Aldrich) were added in order to achieve optic density reading in a spectrometer at al 540nm wave length.
Cell migration assessmentWith cells in confluence, a lesion was performed along the culture, causing continuity loss, the culture was monitored with photographs taken at 24, 48 and 72hours.
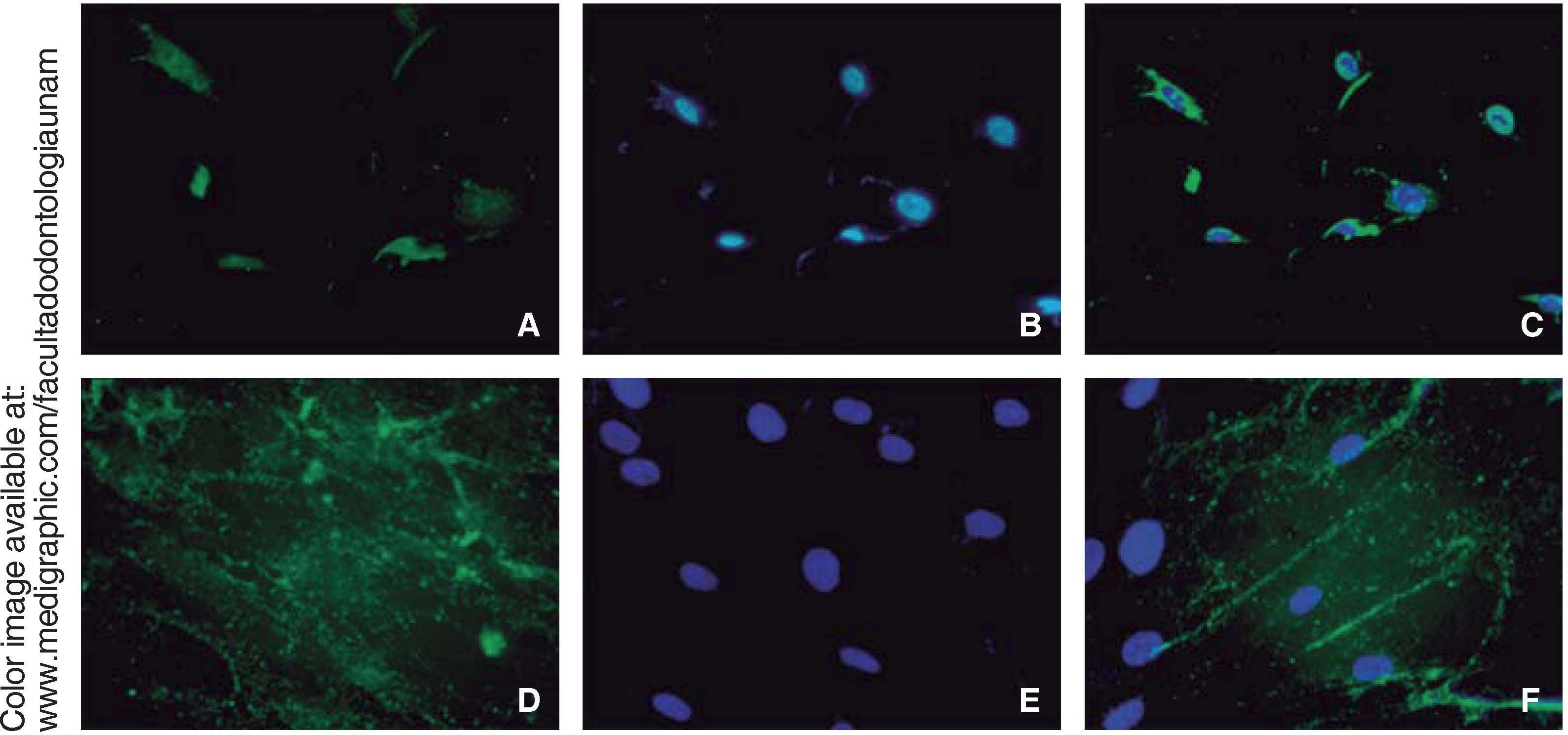
ImmunocytofluorescenceCells were fixated in a 4% paraformaldehyde phosphate tampon for 20minutes. Blocking was conducted in PBS with 3% bovine serum albumin and 0.01% Triton-X 100. Cells were stained with the following: anti-human rabbit antibodies conjugated with Fluor® 647: PE, TRIT, FITC, and DAP (Fluoroshield®), RUNX2, osteocalcin, CD73, CD90, CD105 and vimentin (1:250 Santa Cruz) during 45minutes. Samples were examined in a Zeiss Axio LAB1 fluorescence microscope.
Osteogenic differentiationIn this process, 105 were seeded in passage 3; medium StemPro Osteogenesis Differentiation Kit (Gibco®) was added; it was replaced every 72hours for 14 days.
Staining with alizarin redOnce the cells were differentiated after 7 and 14 days’ culture, they were fixated and stained with alizarin red. They were then subjected to three five minute rinses with PBS; 500μL 0.1% alizarin red were added (HYCEL®) and were left to rest for 15minutes at room temperature.
RESULTSCell population harvestingPoor technical specificity of found articles influenced in obtaining cell population harvesting, cultures conducted in triplicate with each of the studied techniques lead us to obtain a minimum cell concentration which did not achieve confluence. In some cases, after a culture time of seven days, no cells were obtained; and in other cases, the scarce amount of obtained cells was susceptible to contamination, probably caused by the transport medium of the tooth and the washes.
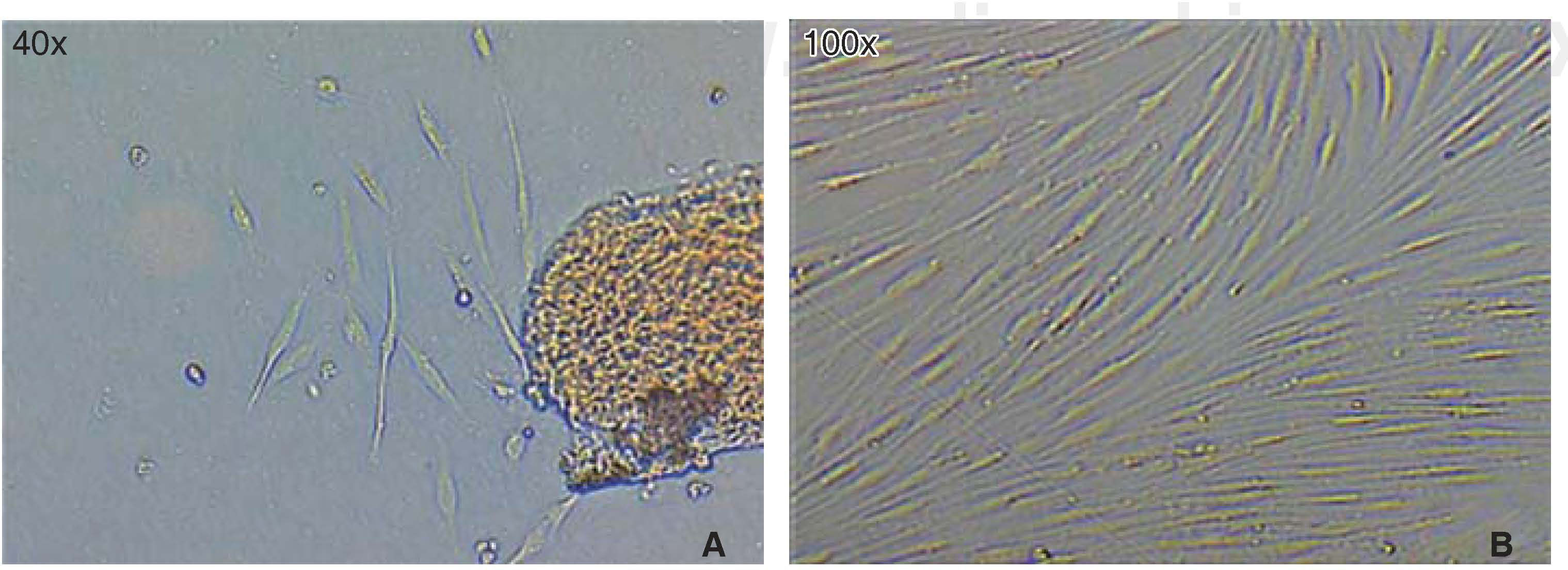
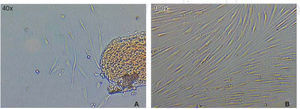
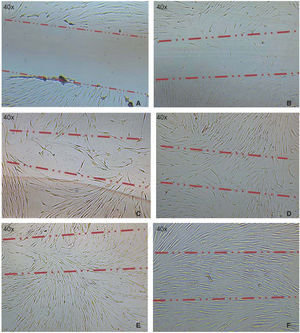
Optimum cell population for the study was achieved using the «standardized technique of enzymatic digestion and explant culture». Once the periodontal tissue was dissected and seeded, cells appeared in the first 72hours, which after an additional period of 46hours showed fibroblastoid aspect in the cultured explants periphery, filling thus the culture dish in 65 to 70%. After this, explants were eliminated with the washes. After seven days, the culture dish was found to exhibit cells in confluence (Figure 1).
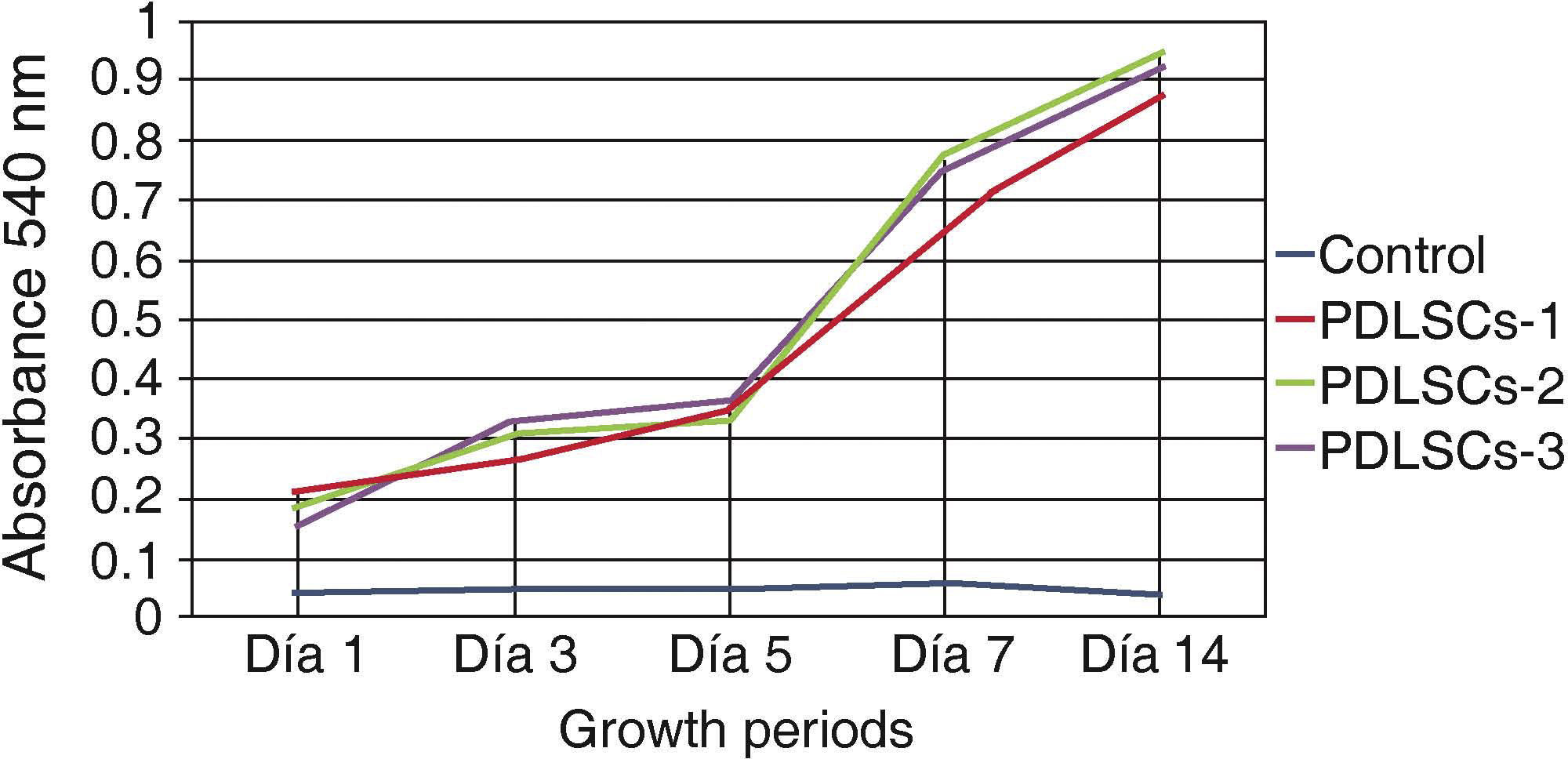
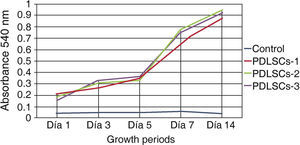
Viability and/or cell proliferationCell population self-renovation capacity was assessed by means of methylthiazole tetrazolium assay (MTT) 3-(4,5-dimethylthiazole 2-il)-2,5-diphenyltetrazolium bromide; spectometry-measured absorbance was proportionate to cell proliferation viability.
Five hundred cells were seeded in 24-well plates; they were evaluated at 1, 3, 5, 7 and 14 days. Absorbance was measured at a 540nm wave length. Obtained measurements were incorporated in a graph. An ascending graph was obtained, which corresponded to a concentration of cells per measuring unit, thus demonstrating that cells preserved their proliferation during 14 days in standard culture circumstances (Figure 2).
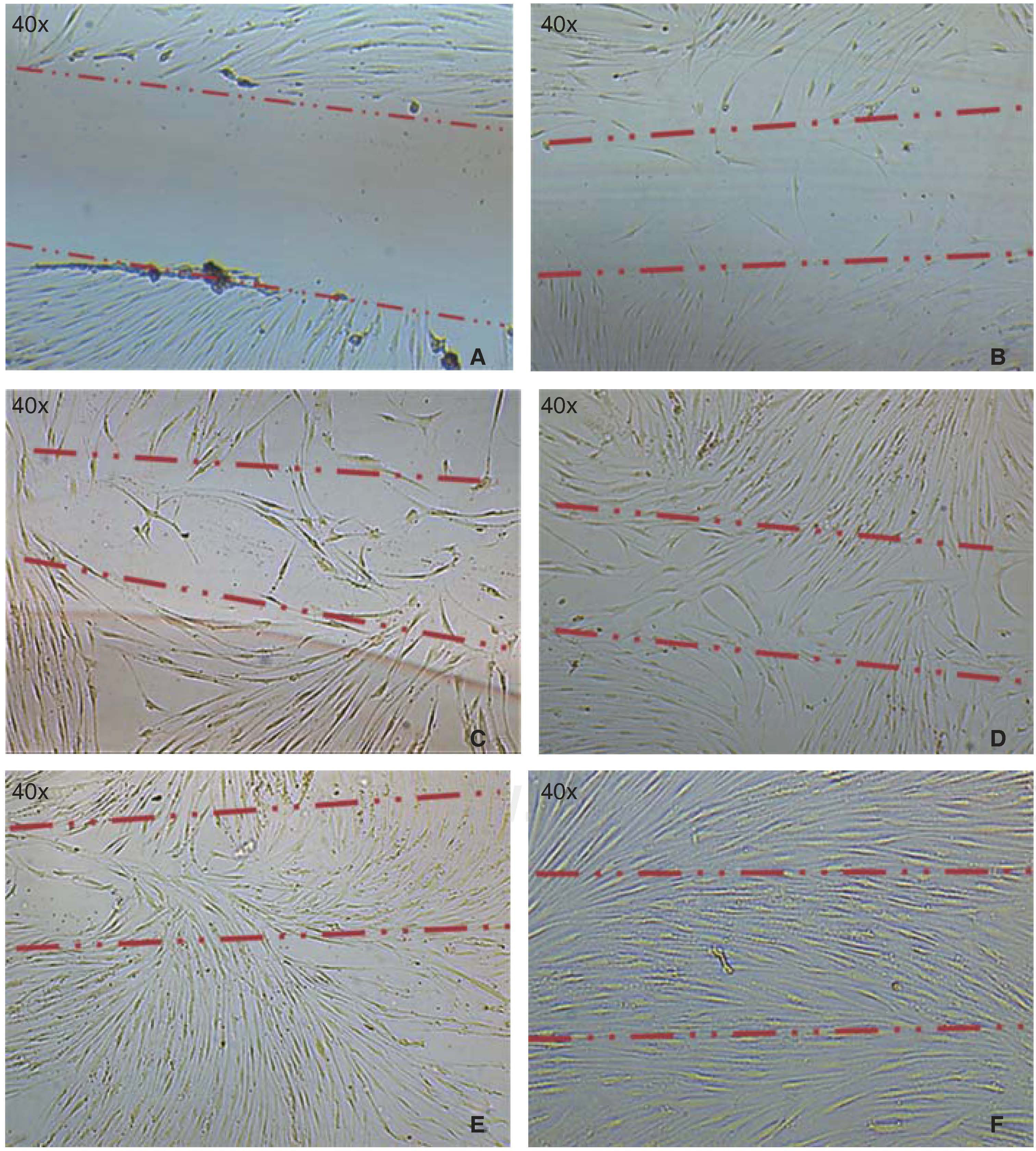
Cell migration capacityCell migration towards the injured area was observed after 12hours of lesion onset; cells coated between 10-20% of the critical area. After 24hours, cell numbers increased, covering then from 40-60% of the surface of the culture jar. After 48hours, cells were observed to be in semi-confluence; after 72hours, cells totally covered the area, this can be observed in images of figure 3.
A) Photograph taken immediately after establishing lesion in the culture. B) 12hours after the lesion, migration of a group of cells re-colonizing 10 to 20% of the lesion can be observed. C) 24hours later migration of a cell group was observed which re-colonized damaged area in 40/60%. D) and E) After 48hours, it can be observed that cells have covered 80-90%. F) 72hours after the lesion, cells reach confluence once again.
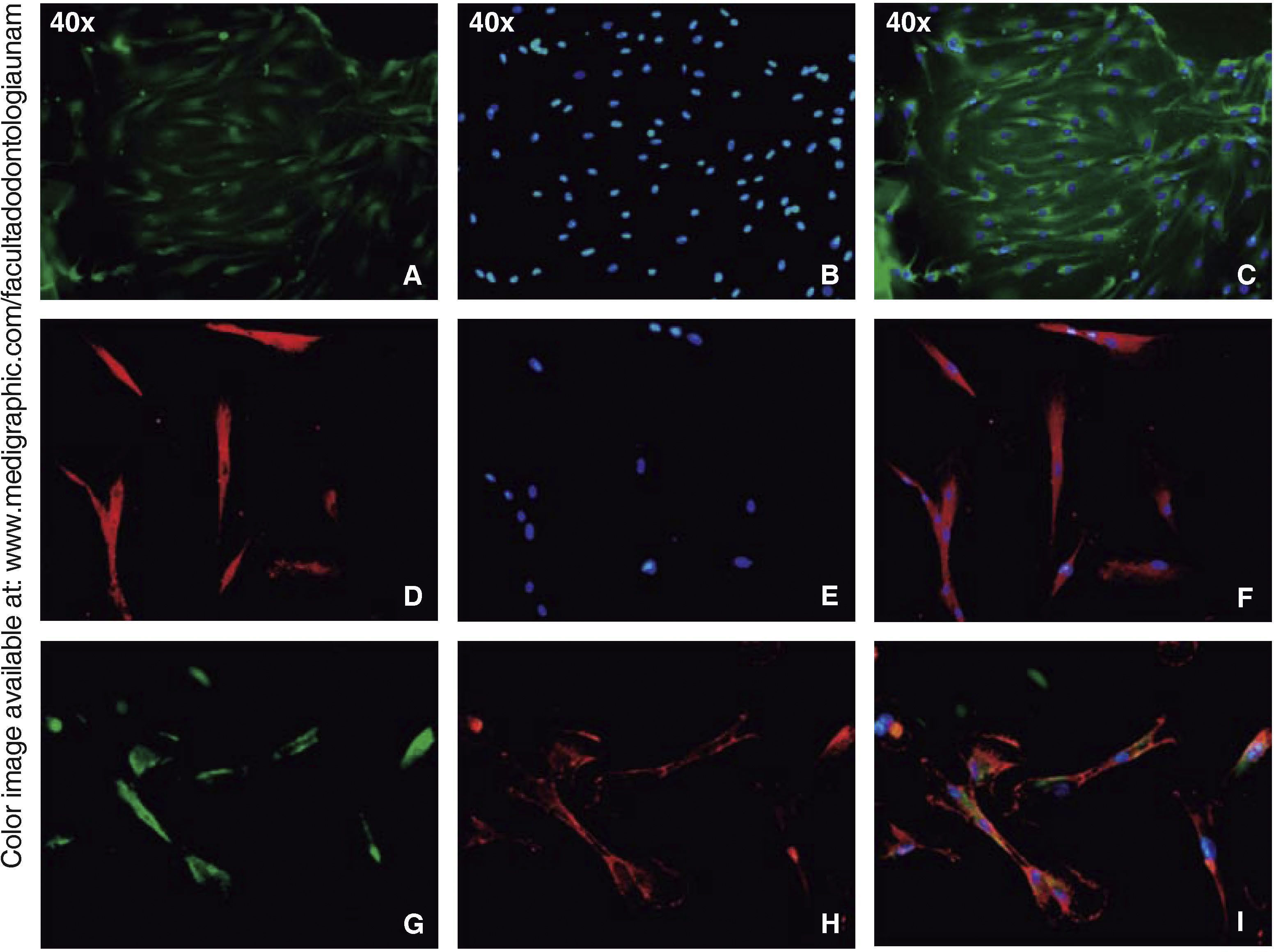
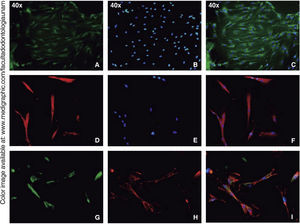
Immunofluorescence assays were performed in the third subculture of our cell population. After counting, positive expression to CD90 was observed in over 80% of all cells, positive expression to CD73 was observed in 70%, positive expression to CD105 in 60% and vimentin in 90% of the population (Figure 4).
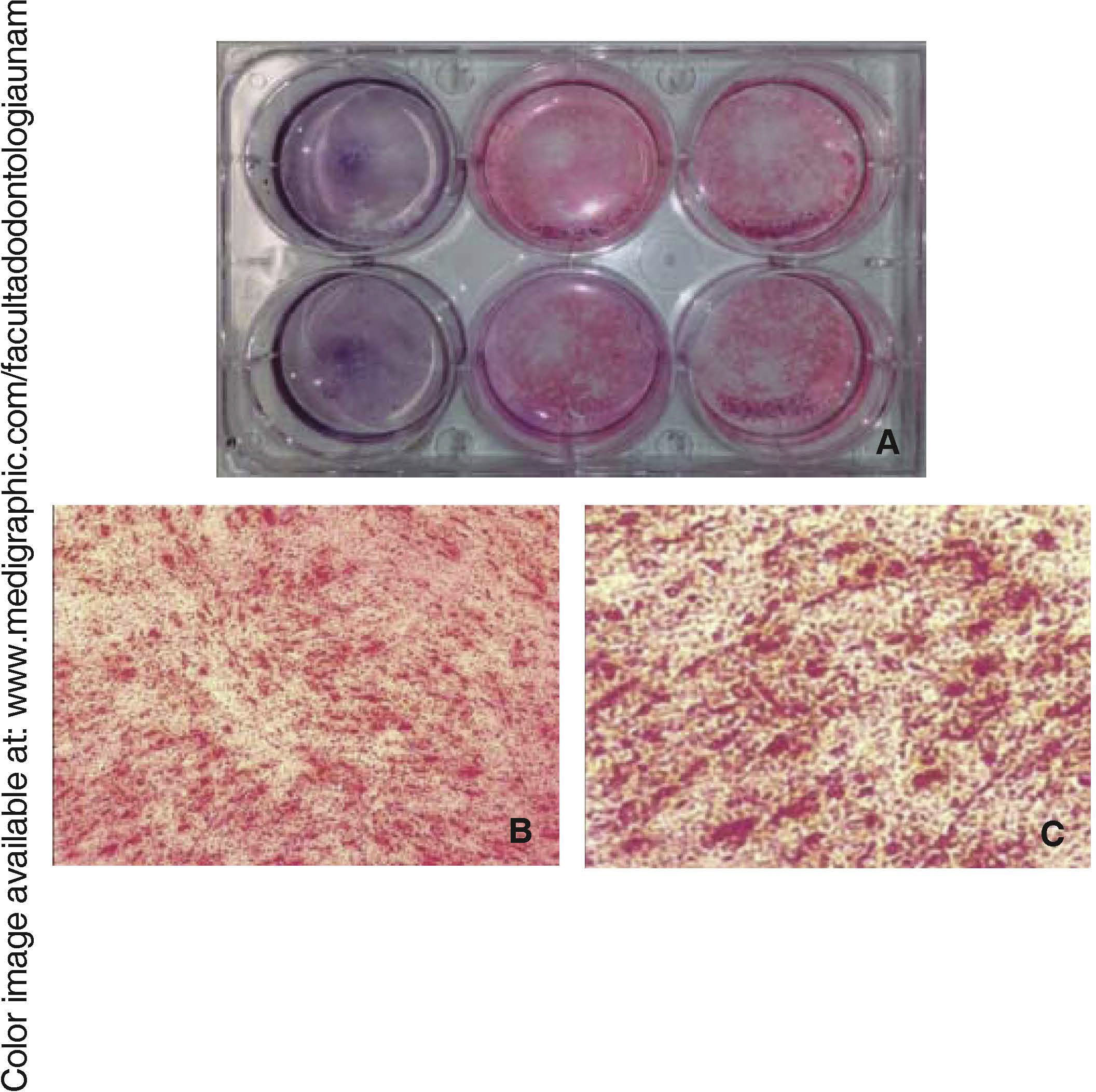
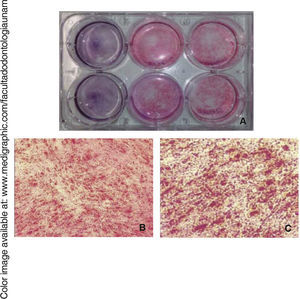
Osteogenic differentiationOnce bone differentiation was induced, cells lost their fibroblastoid morphology in seven days, and acquired a less elongated, cuboid-like prismatic morphology. There was also a greater amount of extracellular matrix as well as presence of calcification nodules, which were observed by means of staining with alizarin red the cultured area, revealing thus a size of 9.6cm2(Figure 5).
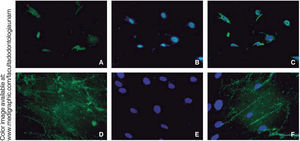
RUNX2 is a transcription factor indispensable for bone maturation, it was expressed in our cell population in osteogenic differentiation at 14 days, in at least 15% of all cells.
Osteocalcin is a protein secreted at late stages of bone differentiation, it is able to bond to calcium for its stabilization. The presence of this molecule in 90% of cells after 14 days indicates mineralization regulation (Figure 6).
DISCUSSIONPeriodontal ligament can be considered an accessible source, since teeth, once extracted, are considered waste tissue, and in cases under orthodontic treatment, extraction is the best treatment alternative.12
It has already been shown that cell populations obtained from the Periodontal Ligament possess the ability to regenerate several tissues,13–16 recently, information has been published on the use of periodontal ligament as a source of stem cells. Nevertheless, a consensus should be reached on used protocols, since these protocols can be very inconsistent and prevent clear comparison of obtained cell populations.17 Therefore, the fact of being familiarized in depth with the mechanism of used techniques, allowed us to suggest this «standardized method», which stressed the importance of preserving the sample's integrity as well as avoiding its contamination.18–20 This culture method requires less experience in periodontal tissue handling, therefore it is easy to achieve and might thus avoid possible culture contamination.
The capacity of our cells derived from PDL to form groups of adhering clonogenic cells with characteristics similar to fibroblasts has been demonstrated with the formation of 1 x 106 cells generated from 1 x 103, cultured at low density in a seven day culture time. This population of cell forming colonies, which we have named PDLSCs, exhibited high rate of proliferation by MTT, similar to the rate observed by other authors.3,8
We thus showed that the new method had improved, or at least was effective in extracting a population of cells similar to periodontal ligament mesenchymal cells which showed positive antigenic profile to mesenchymal cell markers CD105, CD90, CD73 and vimentin, this would confirm their mesodermal origin and mesenchymal phase, and allows us to report their mesenchymal stem cell status.13–15,21 PDLSCs showed that osteogenic line can be differentiated in eight days or less; this might be due to ectomesenchymal origin and concurs with reports of Cha et al22 which report a cell population which differentiates expressing osteogenic markers such as RUNX2, OCN and the formation of mineralization nodules.15
CONCLUSIONSBased on obtained results, we can propose that the simplified method allows isolation of a population of mesenchymal cells derived from periodontal ligament of healthy extracted premolars.
DDS, Dental Research Laboratory Almaraz/UBIMED.
This article can be read in its full version in the following page: http://www.medigraphic.com/facultadodontologiaunam