Currently, the lack of reliable quantitative methods have led different research lines to find a model that predicts the post-mortem interval (PMI). The thanatomicrobiome, present from the moment of death, has been shown to change in predictable ways, allowing a correlation with PMI.
Materials and methodsIn this study, the shifts of the thanatomicrobiome in the region of the posterior small intestine and the ascending colon in Mus musculus during the first 24 h of decomposition have been analysed experimentally. For this purpose, a molecular approach based on the analysis of the 16S ribosomal gene (16S rRNA) and a denaturing gradient gel electrophoresis (DGGE) was adopted, followed by analyses of the ecological diversity indices Alpha and beta diversity.
ResultsThe results based on the analysis of the ecological diversity indices reflected statistically significant changes before 12 h, and a decrease in diversity after 12 h post mortem, this being statistically significant in the two intestinal regions analysed. Moreover, the comparative study of microbial communities indicated distinct and structured changes from the moment of death, with shifts in the degree of similarity from the composition detected in life (PMI 0 h).
DiscussionThese results agree with other studies demonstrating a decrease in microbial diversity. However, under the conditions of the study, this decrease does not begin until 12 h after death.
ConclusionsIn conclusion, by examining the dynamics of bacterial diversity, our study has identified phases during decomposition that could help to improve microbial correlation models for PMI estimation.
En la actualidad, la falta de métodos cuantitativos fiables ha llevado a distintas líneas de investigación a buscar un modelo que prediga el intervalo postmortem (IPM). El tanatomicrobioma, presente desde el momento de la muerte, parece sufrir cambios predecibles y que siguen una correlación con el IPM.
Materiales y métodosSe ha analizado experimentalmente el comportamiento del tanatomicrobioma en la región del intestino delgado posterior y del colon ascendente durante las primeras 24 horas de descomposición en Mus musculus. Para ello, se ha llevado a cabo una aproximación molecular basada en el análisis del gen ribosomal 16S (ARNr 16S) mediante electroforesis en gel con gradiente desnaturalizante (DGGE) y, seguidamente un análisis de la alfa y beta diversidad.
ResultadosLos resultados basados en el análisis de los índices de diversidad ecológica reflejaron cambios estadísticamente significativos antes de las 12 horas, y un descenso de la diversidad a partir de esas 12 horas postmortem, siendo este estadísticamente significativo en las dos regiones intestinales analizadas. Por otro lado, el estudio comparativo de las comunidades microbianas mostró que cambian estructurada y diferenciablemente desde el momento de la muerte, alejándose en similitud de las mostradas en vida (IPM 0 horas).
DiscusiónEstos resultados coinciden con el descenso de la diversidad sugerido a largo plazo por distintos autores. Sin embargo, en las condiciones del estudio se ha visto que este descenso no se inicia hasta las 12 horas.
ConclusiónComo conclusión, se ha podido establecer según los cambios en la diversidad bacteriana, fases de la dinámica bacteriana durante la descomposición que podrían ayudar a mejorar modelos de correlación microbiana para la estimación del IPM.
Death triggers a complex, ecological and dynamic process in organisms, which is known as decomposition. This commences with two other fundamental processes: autolysis and putrefaction.1 Autolysis is one of the first steps in decomposition,2 in which fermentation in cells under anaerobic conditions is insufficient to maintain the membrane gradient. The enzymes contained in cellular compartments are therefore freed into the cytoplasm. These enzymes cause lysis of the cellular membranes, freeing their components into the surrounding tissues. These components may be rich in nutrients such as glycides, amino acids, minerals and water.3 On the other hand, after death the immune system-mediated mechanisms of controlling bacterial proliferation disappear, as do other physical barriers, thereby allowing the commensal bacteria to metabolise these nutrients and proliferate, reaching organs which are sterile under normal conditions (such as the heart, brain, liver or spleen). Nevertheless, some organs are now known to have a resident microbiota, as is the case for the lungs.4 In other cases, sterile organs such as the placenta may cease to be so during infectious processes.5 These microorganisms, whether they are habitually present or are so by chance, start to colonise and disperse through the tissues in a process known as putrefaction. The commensal microbiota, which is present from the moment of death, is a clear focus for the propagation of microorganisms,6 and due to the dysbiosis it undergoes, it is renamed the thanatomicrobiome.7,8
The thanatomicrobiome is a set of bacterial communities which remain isolated and stable during early decomposition. They are therefore less distorted by biotic and abiotic factors than the epinecrotic communities.9 Until fairly recently, knowledge about the composition, successions and interactions between microorganisms was limited by the few tools made available using microbiological culture techniques. Nevertheless, after the 1990s with the emergence of molecular methods, or later at the start of the 21st century with the appearance of massive new generation sequencing (NGS) knowledge has advanced and identified a higher proportion of the microbial species which are involved. The most widely used molecular techniques for analysing microbial communities are based on the analysis of electrophoretic band patterns, such as RFLP, RAPD or denaturing gradient gel electrophoresis (DGGE). The latter technique entered its golden age after 1998, when Muyzer et al.10 described it for the study of microbial communities, including populations that it had been impossible to culture beforehand. It went on to be a method that was used and mentioned in more than 14,000 scientific publications (data obtained from the Pubmed database, www.pubmed.ncbi.nlm.nih.gov, on 19 July 2021). Nowadays, although it has been superseded by NGS techniques, the DGGE technique is still used to study microbial communities quickly, simply and more economically.11–13
NGS techniques have given rise to new disciplines and concepts such as metagenomics, which have made it possible to undertake more precise and sensitive studies. This progress has occurred parallel to the rapid development of high performance sequencing technologies. This in turn has led to increased automation and a reduction in costs, as well as a large amount of information. This technology was used by Metcalf et al.14 in their research into the thanatomicrobiome. These authors were the first to point out that the changes in bacterial communities are predictable in controlled environments and may be used as a biological clock so that they would be of use in estimating the PMI. Although the microbiota varies inter- and intra-individually, the composition of the human microbiota is relatively stable. The intestinal microbiota has been examined in live subjects, and there are what is now known as three enterotypes or differing proportions of the three most abundant microbial groups which are present in the microbiota.15 The characterisation of enterotypes has been very useful in establishing the processes involved in obesity and other diseases. However, the environmental conditions in organisms change after death, so that the communities change, too. This fact has been pointed out by other authors, who mention the existence of species which participate the most in the decomposition process. These species do not vary depending on host variables such as sex, weight or cause of death (for example, metastatic cancer, cardiovascular infarct, cerebral pneumonia or pulmonary obstruction).16 Moreover, thanks to a bioinformatics approach using automatic learning techniques, it is possible to use variations in the thanatomicrobiome when different variables are entered, to find a model of fit that correlates with the PMI.17 The majority of studies undertaken in the past centred on the long-term evolution of the thanatomicrobiome, and in the majority of them the first post mortem measurement analysed took place at 24 h.14,16,18–20
The possibility of cadaveric dating and the lack of a modern quantitative methodology for the estimation of the PMI led to concentrating analysis on the study of the thanatomicrobiome. Additionally, it is present from the moment of death and therefore suffers no delay in colonization, as occurs with insects. Nor are the intestinal communities affected by changes in the location of remains during the early phases of decomposition.7 This study aims to support and add bibliography on the correlation suggested by different authors with the PMI, while also supplying information about the first 24 h to be taken into account in the search for and perfecting of a reliable fit model for estimation of the PMI. This is why a mouse (Mus musculus) model of animal decomposition is used, and due to the lack of bibliography regarding the initial phases of decomposition, the aim is to evaluate the early evolution of the intestinal thanatomicrobiome in the most distal regions of the digestive tract, centring on those close to the caecum. These regions initially have a greater abundance of bacteria21 and are less influenced by temperature,22 which suggests that they may be of interest to obtain a better fitting model for a possible correlation with the PMI.20
Materials and methodsExperimental animals and procedureA total of 25 examples of Mus musculus were used in the experiment, of different ages and sex 1:1 of the endogamic strain C57BL/6. They were fed with a safe diet (ds-A04) and received no treatment. These animals were supplied by the Centre for Animal Experimentation of Malaga University and already sacrificed by exposure to CO2. The moment of sacrifice was established as the starting time for the PMI (0 h). After euthanasia the animals were placed on separate trays, depending on the PMI that had been assigned for their analysis. There were a total of five biological replicas for each time interval (0, 4, 8, 12 and 24 h). The examples were weighed on digital scales, giving an average weight of 36.3 ± 6.7 g. The decomposition process took place at an ambient temperature of 21 ± 3 °C. Once the set PMI had transpired work took place under aseptic environmental and manipulation conditions, using sterile material, a burner and personal protective equipment. To ensure the absence of crossed contamination, an open plate of bacteriological culture was exposed next to the mice (Luria-Bertani, Oxoid, Ireland) during the whole procedure with the animal and sample taking. This plate was subsequently closed and left to incubate for 24–48 h at ambient temperature, before checking the absence of bacterial growth.
After opening the abdominal cavity a distal portion of the posterior small intestine (IC) was extracted, together with a portion of the region of the ascending colon (AC) at the established PMI. Subsequently 500 μL of resuspension buffer was added (0.1 M Tris-HC1; 0.01 M NaCl; 0.1 M EDTA; pH 8.0)23 and they were stored at -20 °C until they were processed.
Molecular analysis of the intestinal thanatomicrobiomeThe samples were gradually unfrozen in ice and were broken down by maceration to free the thanatomicrobiota of the internal surface of the intestine. The mixture was then levelled with a resuspension buffer up to 1 mL. The samples were then homogenised and DNA was extracted according to a standard saline precipitation protocol described previously by Martínez et al.23 with some modifications.24 The DNA was quantified using fluorometric techniques (Qubit v 3.0) and it reached a final concentration of 3 ng/μL with free sterile DNasas water. Lastly, 3 μL of stock DNA was extracted and mixed with 17 μL of free sterile DNasas water and the ARNr 16S fragments were amplified by PCR. 5 μL of Taq polymerase Quanta-bio AccuStart™ PCR ThoughMix® was used. For this purpose, primers designed for the V1–V3 regions of the DNAr 16S were used, with the cycles and conditions described by Tapia-Paniagua et al.24 As a positive control 2 μL of DNA extracted from a pure culture of a Vibrio harveyi strain was added (while as a negative control 2 μl of sterile Milliq water was added). The samples with amplification that did not show intensity at the same level as that obtained in the positive control were excluded for subsequent analysis.
The resulting amplicons were separated and analysed by DGGE as described by Tapia-Paniagua et al.24 DGGE is a widely used molecular technique which in a simple and relatively fast way makes it possible to establish differences between the communities present in two different ecosystems. It is based on the amount or percentage of cytosine–guanine present in the amplified sequences of the universal bacterial marker gene DNAr 16S. The amplified sequences are loaded into DCodeTM System (Bio-Rad Laboratories, Hercules, AC, USA) equipment with a denaturing gradient of from 30% to 55% of urea and formamide, this gradient having been established to discriminate the majority of microbial communities resident in the intestine.24 Electrophoresis was performed at 120 V during 10 min. and at 80 V during 18 h at a constant temperature of 60 °C. Wells were reserved to load 5 μL of 100 pb BioLine HyperLadderTM molecular weight marker. This band weight marker was used to normalise between gels and to homogenise band height based on the height of the bands. The gels were stained using silver stain according to the protocol of Sanguinetti et al.25 and they were scanned with a Bio-Rad, ChemiDoc™ XRS + Molecular Imager® system at a minimum resolution of 300 dpi for subsequent image analysis. The image processing and manual marker band analysis was undertaken using the GelAnalyzer v19.1 programme (www.GelAnalyzer.com). This programme automatically detects the intensity, thickness and positon signals of each one of the marked bands in each sample, giving it a numerical value. Finally, a data intensity matrix is obtained, as well as the presence or absence of bands in each one of the positions which will be subsequently analysed.
Image analysis and thanatomicrobiome diversity studyThe data matrix obtained from the band weight marker was used to calculate the ecological indices according to the criteria of Muyzer et al.10 The richness of species (R) was used to study the ecological indices based on alpha diversity. This consists of the number of different species, based on the number of bands present in each sample subjected to DGGE (R = ∑b, where b is the number of bands present in each lane of the gel). Diversity according to Shannon’s Index (H’)26 refers to the equal share of individuals between the different species (H’ = − ∑Pi ln Pi, where Pi is the intensity of each band divided by the total intensities of all of the bands).To calculate this index, the number of bands was taken into account together with the intensity and thickness of each one of them. The values may stand at from 0 to 5, where 5 is the value expressing maximum diversity. Dominance according to Simpson’s Index (D),27 considered as the inverse of diversity (D = ∑ni (ni-1)/N (N-1)) and, lastly, habitability (Rr) according to the formula described by Marzorati et al.,28 which takes into account the capacity of an ecosystem to hold different species. This is based on the denaturing gradient range in which the bands of each lane are concentrated (Rr = N2 Dg). Values below 30 indicate low habitability, while values higher than 60 are considered to represent highly habitable ecosystems. Lastly, the study of beta diversity is undertaken using a comparative study of microbial communities by means of conglomerate analysis based on Bray-Curtis distance methodology and a graphic representation in dendrogrammes using the UPGMA (Unweighted Pair Group Method with Arithmetic mean) algorithm. The PAST v4.03 (PAleontological STatistics, www.palaeo-electronica.org) programme was used to obtain the ecological indexes and the study of beta diversity.
All statistical tests were undertaken using Statgraphics Centurion XVIII (www.statgraphics.net) offline software. To validate the tests of normalcy and homoscedasticity a parametric study was conducted by means of variance analysis (ANOVA), in this case with multiple range tests, grouping data in homogeneous sets; according to Fisher’s LSD (Least Significant Difference) test, belonging to different groups indicated that a significant difference existed (P <.05).
ResultsAlpha diversity analysisThe ecological indices used to study alpha diversity in the posterior region of the IC and the region of the AC are shown in Table 1; both sets of data were validated in the normalcy and homoscedasticity tests. The values of the ecological indices in the IC region remained stables, without showing significant differences until 12 h. PMI in the R, H’ and in the Rr, while D increased significantly at PMI 12 h. In the AC region, the values of the indices calculated increased significantly until PMI 12 h from the moment of death, except for Rr. Finally, in both regions the values of all of the above-mentioned ecological indices fall significantly at PMI 24 h, and all of them at PMI 24 h were statistically inferior to those obtained beforehand at PMI 12 h.
Species richness values (R), dominance (D), diversity (H’) and habitability (Rr) obtained based on analysis of bacterial communities in the posterior region of the small intestine (IC) and in the region of the ascending colon (AC), in post-mortem examples of Mus musculus at different known intervals (PMI), shown here in hours. The values of the ecological indices are shown as the average values of the replicas ± standard error. Different letters in the same column denote significant differences (P < .05) from the other PMI values for the same parameter.
| PMI (h) | R | D | H’ | Rr |
|---|---|---|---|---|
| Posterior region of the small intestine (IC) | ||||
| 0 | 17.7 ± 1.6a | .918 ± .007ac | 2.70 ± .09ab | 78.48 ± 14.16a |
| 4 | 15.8 ± 1.3ab | .929 ± .006abc | 2.70 ± .08ab | 59.66 ± 12.27ab |
| 8 | 17.5 ± 1.3a | .933 ± .006ab | 2.78 ± .08b | 72.97 ± 12.27a |
| 12 | 19.4 ± 1.2a | .941 ± .006b | 2.89 ± .07b | 90.76 ± 10.97a |
| 24 | 13.0 ± 1.2b | .915 ± 0.006c | 2.51 ± .07a | 28.37 ± 10.97b |
| Region of the ascending colon (AC) | ||||
| 0 | 13.5 ± 1.2ab | .897 ± 0.006a | 2.44 ± .08a | 45.84 ± 8.90ab |
| 4 | 16.3 ± 1.2bc | .927 ± 0.006bc | 2.70 ± .08bc | 64.77 ± 8.90b |
| 8 | 17.0 ± 1.1c | .937 ± 0.006cd | 2.80 ± .07c | 63.57 ± 7.96b |
| 12 | 22.2 ± 1.1d | .951 ± 0.006d | 3.06 ± .07d | 116.14 ± 7.96c |
| 24 | 12.8 ± 1.1a | .914 ± 0.006ab | 2.50 ± .07ab | 27.39 ± 7.96a |
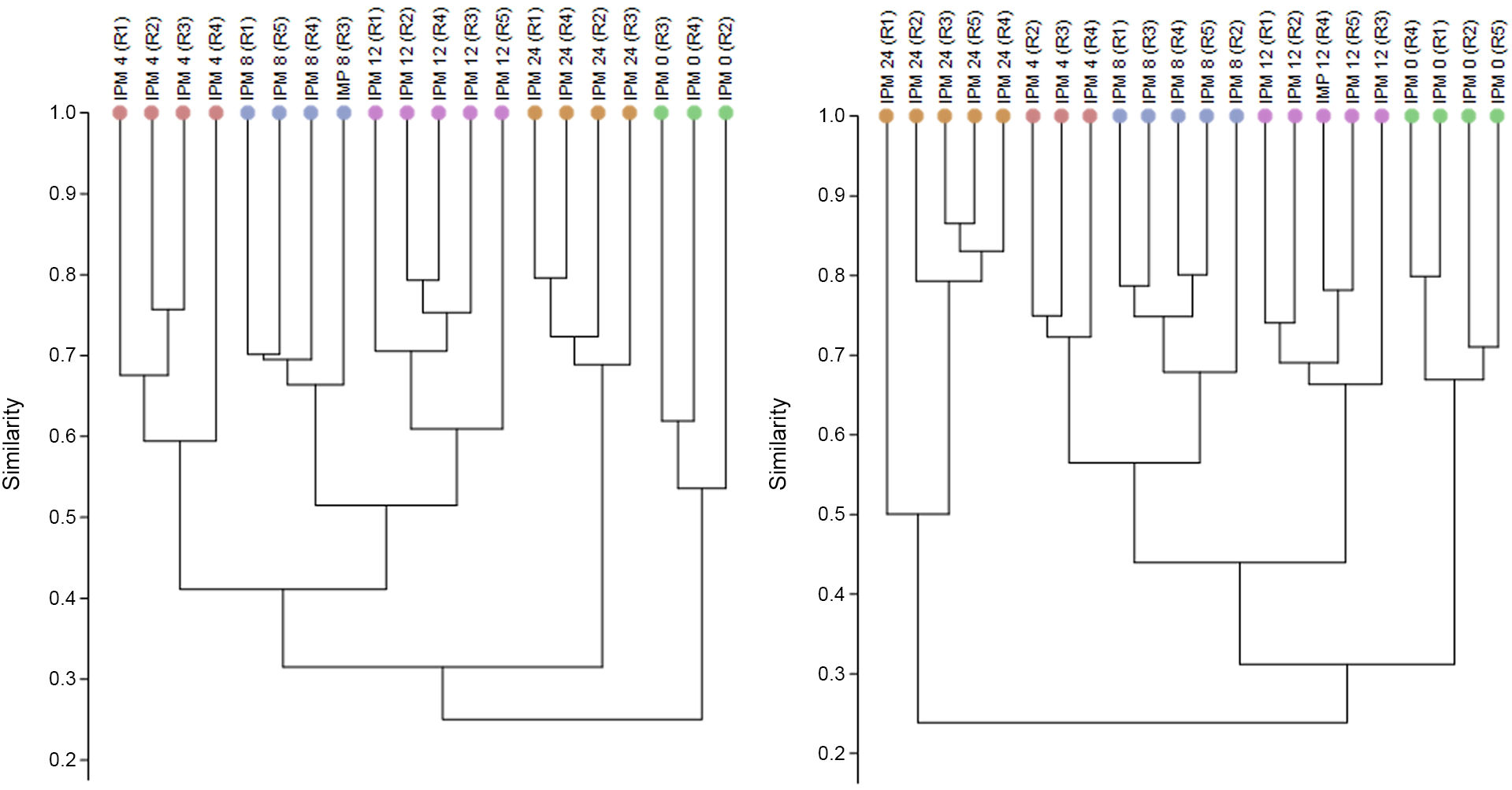
The dendrogrammes obtained based on image analysis of the DGGE gels (for example, supplementary material) are shown in Fig. 1. The dendrogramme is created on the basis of the Bray-Curtis index, defining the value of 0 as the minimum similarity or maximum dissimilarity, and 1 as samples which have identical microbial communities. The similarity study showed that the bacterial communities were grouped according to the PMI they corresponded to, thereby differentiating them from the others; this occurred in the IC as well as in the AC region. In both regions studied, the bacterial communities at PMI of 4 h, 8 h and 12 h had a compositional similarity of around 0.4–0.5, or equally, in terms of percentage of similarity, of 40%–45%. Moreover, in the regions studied the bacterial communities which were the least similar attained values of from 0.2 to 0.3 (around 20%–30%) and corresponded to PMI 0 h as opposed to those at 24 h. On the other hand, the biological replicas at the same PMI had a similarity with values that run from 0.5 to 0.7, which seems to indicate that there is at least 30% of variation in the composition of the microbial communities between examples within a single PMI, or what is known as intrinsic individual variability.
Dendrogrammes obtained using the Bray-Curtis distance method based on analysis of the intestinal bacterial communities of Mus musculus examples at different known post-mortem intervals (PMIs) shown in hours; a single colour represents a single PMI. Each dendrogramme represents a different intestinal region studied, these being the posterior small intestine (IC) and the ascending colon (AC). The biological replica in question is shown in brackets (Rn°) at a specific PMI.
A total of 50 intestinal samples were analysed using molecular techniques based on ARNr 16S analysis, obtaining a molecular approximation of the thanatomicrobiome. The results obtained in this study reveal information on the behaviour of the thanatomicrobiome during the early phases of the decomposition process in a mouse animal model. In the study of alpha diversity, significant changes were found in the bacterial communities analysed in both intestinal regions during the first hours of decomposition. The Simpson dominance values (D) revealed a high dominance during all of the PMI in both intestinal regions, with similar values to those shown at PMI 0 hrs. and 24 h in other murine studies using sequencing techniques.19 This may be expected due to the environmental specificity of the intestines from an ecological point of view,21 as the majority of intestinal bacteria in mice at the moment of death belong to the phyla Firmicutes, Bacteriodedes and Proteobacteria.14 On the other hand, in all of the intestinal regions the Shannon diversity values (H’) remained within a range of intermediate diversity in the PMI studied, and these results agree with those of other authors,20 who also found a diversity defined as intermediate in the initial stages of decomposition. The analysis by Liu et al.20 was able to classify all of their sequenced taxonomic units in 29 phyla, 67 classes, 160 orders, 250 families, 473 genera and 835 bacterial species. One possible explanation of the levels of diversity from an evolutionary viewpoint may be that the bacteria which live in the intestines are specialised and adapted, so that they live in a symbiotic relationship with the host. Lastly, the habitability (Rr) values varied from low habitability up to medium and high habitability at the different PMI during decomposition, reflecting the changes in the intestinal environment during decomposition. The Rr values at a PMI of 24 h showed low habitability in all of the regions, indicating a tendency towards a restrictive environment that would also favour the emergence of more specialised microbial species.
In general, in both intestinal regions the indices which analyse alpha diversity displayed the same tendency from the moment of death until the PMI at 12 h, and this may be due to post-mortem intestinal dysbiosis. Death would cause a period or phase of adaptation in the intestinal bacterial communities to the new state of entropy; the conditions of decomposition and the study model used in this work indicate that this adaptation phase lasts until a PMI of 12 h. Additionally, as the residual oxygen in tissues during decomposition is exhausted, an ecological succession occurs in which aerobic microorganisms are replaced by anaerobic ones.19 The possible hypothesis of a fall in tissue oxygen after death would support the existence of an adaptation phase as proposed in the alpha diversity study. During decomposition the accumulation of gases soon becomes critical and the abdominal cavity opens to the environment. Several studies state that this moment marks a fundamental change in the bacterial communities,14,16 as the opening of the abdominal cavity causes a fall in anaerobic populations due to the entry of oxygen, so that aerobic microorganisms become dominant. According to the study by Metcalf et al.14 in a set of mice in uniform conditions, abdominal opening took place from 6 to 9 days after death, and this makes the fit in abdominal data after 48 days to estimate the PMI less useful. This shows that this event is variable, and in the current study of beta diversity the bacterial communities showed an inter-individual variation of at least 30%, which would support the said temporal variation. Abdominal opening is linked to the decomposition process, and the latter is in turn influenced by abiotic and biotic factors such as bacterial composition.
Alpha diversity subsequently falls after a PMI of 12 h down to significantly lower values at the 24-h PMI. The alpha diversity tendency found in the 24-h PMI agrees with several microbiological studies of decomposition in humans or animal models. These studies show that bacterial communities tend to increase in relative abundance and fall in terms of bacterial diversity and richness in comparison with their initial levels at PMI of 0 h, with the progress of decomposition in a cadaver.14,16,18–20 Lastly, the beta diversity study found that both intestinal regions at each PMI had their own specific bacterial community composition and a profile that could be differentiated from those at the other PMIs. Li et al.,19 in a study of beta diversity in murines by analysis of the main components (PAC) during a long-term decomposition process, showed that bacterial communities are clearly grouped according to their corresponding PMI. In the present study, the limiting PMI (at 0 h and at 24 h) were the ones which differed the most in this respect, while all of the PMI were different from the others, suggesting that the variation in bacterial communities is gradual and follows a pattern. As was suggested by Metcalf et al.,14 the thanatomicrobiome undergoes changes links to the decomposition process, and with time it becomes increasingly unlike the initial communities within the abdominal cavity.
Bacterial composition is linked to the decomposition process, and although decomposing bacterial communities eventually become similar, other studies show that activity varies depending on seasonal factors, chiefly temperature, which is the most important abiotic factor.29 This study based on molecular approximation reflect an adaptation phase lasting for 12 h post mortem. Based on the available bibliography on bacterial behaviour during long-term decomposition, it is suggested that future research into the thanatomicrobiome should centre their model of fit on three study windows, to hopefully increase reliability: 1) until the adaptation phase; 2) from the end of the previous stage to the resolution of bloating; and lastly, or 3) until the cessation of bacterial activity. The fact that these intervals last for varying periods of time must be taken into account. This is due to the decomposition process and, in the case of the last of the said windows, the opening of the cadaver to the environment would lead to the latter have greater influence on the variation in the bacterial communities, so that the same reliability could not be expected, making it less useful. Therefore, estimating the PMI based on variation in the thanatomicrobiome has to take into account the study window in question and the environment. Liu et al.20 made a model of fit in the murine caecum that indicated two study windows: during the first 24 h, obtaining a reliability of 1.5 ± 0.8 h; and up to 15 days, obtaining a reliability of 14.5 ± 4.4 h.
Study of the changes in bacterial diversity has made it possible to establish phases in the bacterial dynamic during decomposition. The adaptation phase, which is similar to the bacterial growth curve in a culture under optimum conditions,30 may be described in this case as the period of bacterial readjustment after death and previous to the fall in diversity. These terms have been suggested and commented on in this microbiological study of decomposition, and they aim to improve the models of fit used to estimate the PMI. Nevertheless, taking its limitations into account and due to the lack of bibliography which centres on the early stages of decomposition, it requires validation by other more precise studies, using massive sequencing, for example. Additionally, studies are also required on the duration of decomposition, as this may vary with the environment and/or species. It also has to be said that this study in animal models has been able to use multiple biological replicas, while to be useful in the forensic sciences it has to be extrapolated to human beings. To conclude, this study centres on early PMIs, and it supports the fall in bacterial diversity during decomposition suggested by other authors, showing that under the conditions analysed this fall commences after 12 h. On the other hand, the beta diversity study showed that bacterial communities are grouped according to the PMI they correspond to, and that they tend to differentiate from those found initially as other studies suggest, which would support the suggested hypothesis of the possible utility of the thanatomicrobiome in estimating the PMI.
FinancingNone.
Conflict of interestsThe authors have no conflict of interests to declare.
The authors would like to thank Ricardo González Carrascosa, veterinary surgeon in charge of the Animal Experimentation Centre of Malaga University, for supplying the animals used in this work.
Please cite this article as Aragonés A, Martínez-Manzanares E, Tapia-Paniagua ST. Estudio mediante técnicas moleculares del tanatomicrobioma intestinal en la estimación del intervalo post mortem temprano empleando un modelo de ratón. Rev Esp Med Legal. 2022. https://doi.org/10.1016/j.remle.2022.02.002.