Allergic rhinitis (AR) is a polygenic inflammatory disorder of the upper respiratory airway with an increasing prevalence worldwide. Interleukin-10 (IL-10) and transforming growth factor-beta (TGF-β), as two cytokines with pleiotropic effects on both innate and adaptive immunity, play important roles in allergic responses. Therefore, this study was performed to evaluate the associations of five polymorphisms of IL-10 and TGF-β genes with AR.
Materials and methodsNinety-eight patients with AR along with 140 healthy volunteers with no history of AR and with the same ethnicity of the patients were recruited in this study. Genotyping was done for three polymorphisms in promoter region of IL-10 gene (rs1800896, rs1800871, rs1800872), and two polymorphisms in the exonic region of TGF-β1 gene (rs1982037, rs1800471) using PCR sequence-specific-primers method.
ResultsA allele and AA genotype in rs1800896 of IL-10 and TT genotype in rs1982037 in TGF-β were significantly less frequent in the patients than in controls. While the C allele and the CG genotype in rs1800471 in TGF-β1 were associated with a higher susceptibility to AR. C/C and T/C haplotypes (rs1982037, rs1800471) in TGF-β1 gene and A/C/A, A/T/C and G/C/A haplotypes (rs1800896, rs1800871, rs1800872) in IL-10 gene were found with higher frequencies in patients than controls. Patients with CC genotype in rs1800871 in Il-10 had significantly lower levels of IgE.
ConclusionWe found that certain genetic variants in IL-10 and TGF-β polymorphisms were associated with susceptibility to AR as well as some clinical parameters in the patients with AR.
Allergic rhinitis, as the most common allergic disorder, is a type 1 hypersensitivity disorder of the upper airway. AR with clinical symptoms such as sneezing, nasal congestion, nasal pruritus, rhinorrhoea, cognitive impairment and sleep disturbance has a significant effect on patients’ social life, school and work productivity.1–3 AR affects more than 600 million people worldwide and is, therefore, a global health problem with an incidence of 10–30% in adults and 10–46% in children, with an increasing pattern in the last 30–40 years.2 In addition, as the concept of “one airway, one disease” implies, AR is a warning sign for other allergic conditions such as asthma and allergic rhinobronchitis.4,5
In recent years, the pathogenesis of AR has been extensively studied, which paved the pathway to understanding the relationship between Th1/Th2 immune response, allergen-specific immunoglobulin production, inhaled allergens and the underlying inflammatory process responsible for symptoms of AR.1,3 However, a strong heritability component of 0.33–0.75 for AR and identification of several susceptibility loci including 1q31–q32, 3q13, 5q33.1, 19q13.2–4 show that behind this scene, there is a complex interplay between genetic and environmental factors.6–9 Genetic polymorphisms in genes of different cytokines, as modulating factors of the inflammatory reactions, are able to change the secretory levels of immune mediators at several levels from expression patterns to post-translation modifications. They therefore have a great influence on the allergic response towards different allergens and consequently susceptibility to allergic diseases like AR.9,10 We have previously found associations between AR and several genetic polymorphisms in interleukin 1 (IL-1), IL-2, IL-4, IL-4R, IL-6, IL-12, tumour necrosis factor (TNF-α), and interferon-gamma (IFN-γ).11–13
Genes encoding for IL-10 and transforming growth factor beta (TGF-β), two of the major cytokines involved in the pathogenesis of AR, are located at 1q31–1q32 and long arm of chromosome 19 (19q13.1), respectively, which is consistent with the previously identified susceptibility loci.6,7,14 Several common variants including −1082A>G (rs1800896), −819C>T (rs1800871), −592A>C (also called −571 or rs1800872) have been identified within the promoter region of IL-10 gene.15 Two of the common variants in the exonic region of TGF-β1 are +29T>C (rs1982037) and +915G>C (rs1800471).16 Previously, genetic alleles in these single nucleotide polymorphisms (SNP) were associated with susceptibility to several inflammatory diseases, including asthma.17–21
With these facts in mind, these common variants in these cytokines are good candidates for a candidate gene approach study. To the best of our knowledge, up to now, no study has investigated the relationship of these polymorphisms in allergic rhinitis patients. Therefore, in this study, we investigated the associations of three SNPs of IL-10 (rs1800896, rs1800871, rs1800872), and two polymorphisms of TGF-β1 gene (rs1982037, rs1800471) with AR.
Material and methodsStudy populationIn this case-control study, we compared allele frequencies of 98 patients with AR with 140 healthy controls. Ninety-eight patients with AR from a children medical centre were enrolled as the patient group with stratified randomisation for gender, severity and intermittency of disease. The diagnosis of AR and the classification of the severity of the disease in patients were established by an allergologist based on the Allergic Rhinitis and its Impact on Asthma (ARIA) document as follow22: (1) mild means that troublesome symptoms and sleep disturbance are not present and daily activities, leisure and/or sport, school or work activities are not impaired. (2) Moderate/severe means that one or more of the following items are present: sleep disturbance, impairment of daily activities, leisure and/or sport, impairment of school or work, presence of troublesome symptoms.
Demographic and clinical data were gathered by interview, in addition to using patients’ medical files. Total serum immunoglobulin (Ig) E levels were measured by ELISA method.
One hundred and forty healthy and ethnically matched volunteers from blood donors at Iranian blood transfusion organisations were recruited as the control group to estimate background population allele frequencies. Individuals with family history of allergic rhinitis were excluded from the investigation.
Informed consents were obtained from all the participants in both groups. All the principles of the Declaration of Helsinki were applied in the study and the study was approved by the ethical committee of Tehran University of Medical Sciences.
SNP genotypingTo determine allele frequencies for three SNPs of IL-10 (−1082A>G; rs1800896), −819C>T; rs1800871, −592A>C; rs1800872, and two polymorphisms of TGF-β1 gene (+29T>C; rs1982037 and +915G>C; rs1800471), genomic DNA was isolated by modified “salting out” method from peripheral blood from all participants. Then genotyping was performed using polymerase chain reaction sequence-specific-primers (PCR-SSP) methods by Heidelberg cytokine gene polymorphism SSP kit (Heidelberg University, Heidelberg, Germany) as previously described.11–13 To visualise PCR products, 2% agarose gel electrophoresis was done on an ultraviolet transilluminator.
Statistical analysisSPPS version 18 software (SPSS Inc., Chicago, IL, USA) was used to perform all statistical analysis. Crude odds ratios (OR) along with their 95% confidence intervals (CI) were calculated by comparing the frequencies of allele and genotype among patients and controls. Comparisons of frequencies for two groups were tested by chi-square tests and Fisher's test. One-way ANOVA test was used in order to find associations between numeric variables such as IgE levels, eosinophil count, and eosinophil percentage and genotypic subgroups. A p-value less than 0.05 was considered statistically significant.
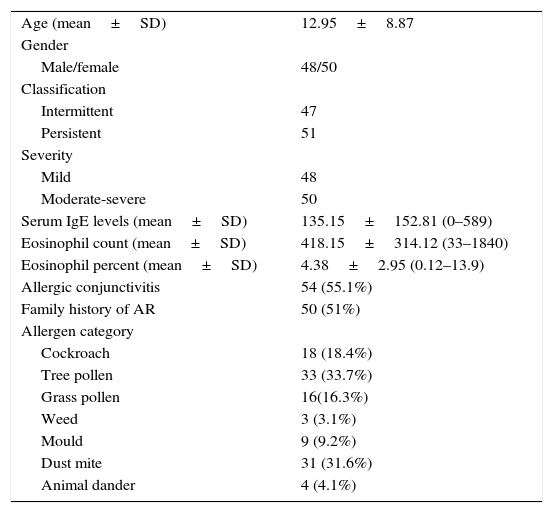
ResultsAt the end of the study, the clinical information and frequencies of five SNPs of IL-10 and TGF-β in 98 patients with AR and three SNPs of IL-10 in 140 healthy individuals and two SNPs of TGF-β in 138 healthy individuals were gathered. The mean age of the patients group was 12.95 (SD=8.87, range=3–44). There were slightly more women than men. Most of the patients were allergic to tree pollen and dust mite (Table 1).
Clinical characteristics of patients with AR.
| Age (mean±SD) | 12.95±8.87 |
| Gender | |
| Male/female | 48/50 |
| Classification | |
| Intermittent | 47 |
| Persistent | 51 |
| Severity | |
| Mild | 48 |
| Moderate-severe | 50 |
| Serum IgE levels (mean±SD) | 135.15±152.81 (0–589) |
| Eosinophil count (mean±SD) | 418.15±314.12 (33–1840) |
| Eosinophil percent (mean±SD) | 4.38±2.95 (0.12–13.9) |
| Allergic conjunctivitis | 54 (55.1%) |
| Family history of AR | 50 (51%) |
| Allergen category | |
| Cockroach | 18 (18.4%) |
| Tree pollen | 33 (33.7%) |
| Grass pollen | 16(16.3%) |
| Weed | 3 (3.1%) |
| Mould | 9 (9.2%) |
| Dust mite | 31 (31.6%) |
| Animal dander | 4 (4.1%) |
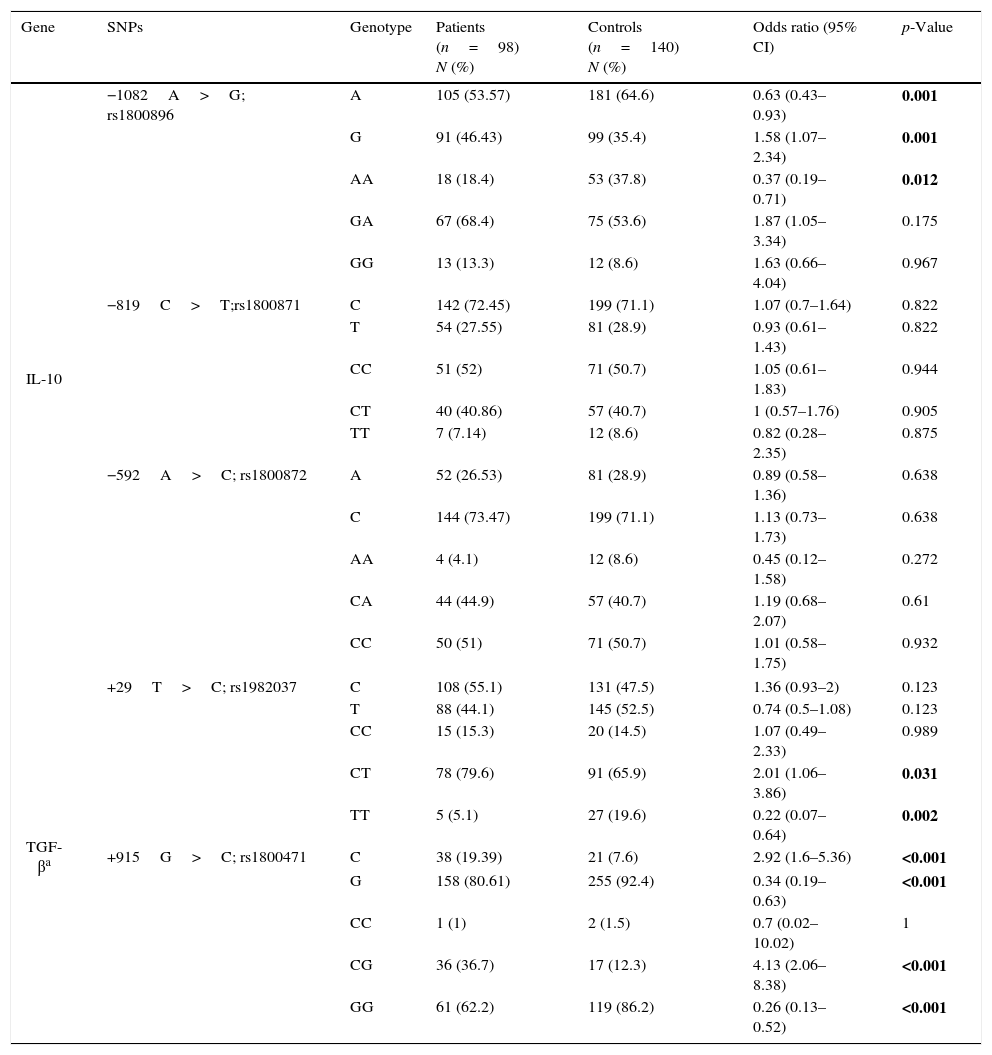
Allele frequencies in the five aforementioned SNPs in IL-10 and TGF-β1 genes were compared between patient and control groups and the odds ratios and the related p-values were computed (Table 2). There was no deviation from Hardy–Weinberg equilibrium for all genotypes in both groups. A allele and AA genotype in rs1800896 of IL-10 were significantly less frequent in the patients than in controls while G allele and GA genotype had significantly higher frequencies in patients. There was not any significant association between allele frequencies of rs1800871 and rs1800872 and susceptibility to AR.
Allele and genotype frequencies in patients and controls.
| Gene | SNPs | Genotype | Patients (n=98) N (%) | Controls (n=140) N (%) | Odds ratio (95% CI) | p-Value |
|---|---|---|---|---|---|---|
| IL-10 | −1082A>G; rs1800896 | A | 105 (53.57) | 181 (64.6) | 0.63 (0.43–0.93) | 0.001 |
| G | 91 (46.43) | 99 (35.4) | 1.58 (1.07–2.34) | 0.001 | ||
| AA | 18 (18.4) | 53 (37.8) | 0.37 (0.19–0.71) | 0.012 | ||
| GA | 67 (68.4) | 75 (53.6) | 1.87 (1.05–3.34) | 0.175 | ||
| GG | 13 (13.3) | 12 (8.6) | 1.63 (0.66–4.04) | 0.967 | ||
| −819C>T;rs1800871 | C | 142 (72.45) | 199 (71.1) | 1.07 (0.7–1.64) | 0.822 | |
| T | 54 (27.55) | 81 (28.9) | 0.93 (0.61–1.43) | 0.822 | ||
| CC | 51 (52) | 71 (50.7) | 1.05 (0.61–1.83) | 0.944 | ||
| CT | 40 (40.86) | 57 (40.7) | 1 (0.57–1.76) | 0.905 | ||
| TT | 7 (7.14) | 12 (8.6) | 0.82 (0.28–2.35) | 0.875 | ||
| −592A>C; rs1800872 | A | 52 (26.53) | 81 (28.9) | 0.89 (0.58–1.36) | 0.638 | |
| C | 144 (73.47) | 199 (71.1) | 1.13 (0.73–1.73) | 0.638 | ||
| AA | 4 (4.1) | 12 (8.6) | 0.45 (0.12–1.58) | 0.272 | ||
| CA | 44 (44.9) | 57 (40.7) | 1.19 (0.68–2.07) | 0.61 | ||
| CC | 50 (51) | 71 (50.7) | 1.01 (0.58–1.75) | 0.932 | ||
| TGF-βa | +29T>C; rs1982037 | C | 108 (55.1) | 131 (47.5) | 1.36 (0.93–2) | 0.123 |
| T | 88 (44.1) | 145 (52.5) | 0.74 (0.5–1.08) | 0.123 | ||
| CC | 15 (15.3) | 20 (14.5) | 1.07 (0.49–2.33) | 0.989 | ||
| CT | 78 (79.6) | 91 (65.9) | 2.01 (1.06–3.86) | 0.031 | ||
| TT | 5 (5.1) | 27 (19.6) | 0.22 (0.07–0.64) | 0.002 | ||
| +915G>C; rs1800471 | C | 38 (19.39) | 21 (7.6) | 2.92 (1.6–5.36) | <0.001 | |
| G | 158 (80.61) | 255 (92.4) | 0.34 (0.19–0.63) | <0.001 | ||
| CC | 1 (1) | 2 (1.5) | 0.7 (0.02–10.02) | 1 | ||
| CG | 36 (36.7) | 17 (12.3) | 4.13 (2.06–8.38) | <0.001 | ||
| GG | 61 (62.2) | 119 (86.2) | 0.26 (0.13–0.52) | <0.001 | ||
Bold values are statistically significant.
Hemizygosity for the T allele of rs1982037 in TGF-β1 was also less frequent in the patients with AR comparing to controls. While the C allele and the CG genotype in rs1800471 in TGF-β1 were associated with a higher susceptibility with AR, GG genotype was less frequent in patients than controls.
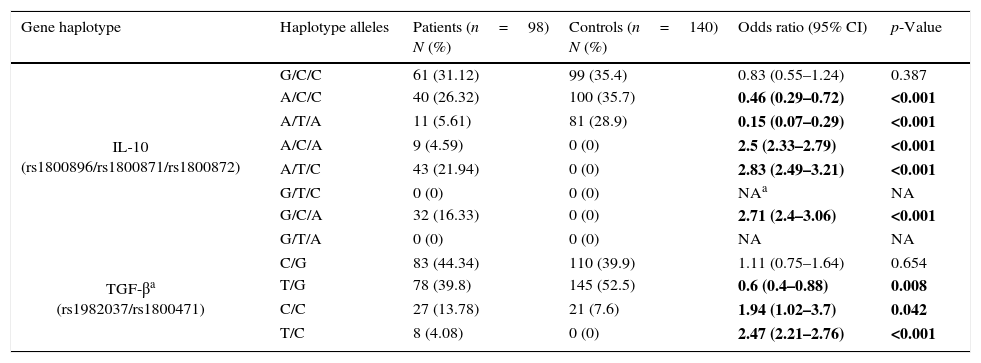
Frequency of haplotypesFrequencies for possible haplotypes in the patients group and healthy controls are also shown in Table 3. Comparison of IL-10 (rs1800896, rs1800871, rs1800872) haplotypes between the patients and controls revealed that frequencies of A/C/A, A/T/C, G/C/A haplotypes were significantly higher in the patient group than in controls while A/T/A and A/C/C were significantly lower in patients.
Haplotype frequencies in patients group and control group.
| Gene haplotype | Haplotype alleles | Patients (n=98) N (%) | Controls (n=140) N (%) | Odds ratio (95% CI) | p-Value |
|---|---|---|---|---|---|
| IL-10 (rs1800896/rs1800871/rs1800872) | G/C/C | 61 (31.12) | 99 (35.4) | 0.83 (0.55–1.24) | 0.387 |
| A/C/C | 40 (26.32) | 100 (35.7) | 0.46 (0.29–0.72) | <0.001 | |
| A/T/A | 11 (5.61) | 81 (28.9) | 0.15 (0.07–0.29) | <0.001 | |
| A/C/A | 9 (4.59) | 0 (0) | 2.5 (2.33–2.79) | <0.001 | |
| A/T/C | 43 (21.94) | 0 (0) | 2.83 (2.49–3.21) | <0.001 | |
| G/T/C | 0 (0) | 0 (0) | NAa | NA | |
| G/C/A | 32 (16.33) | 0 (0) | 2.71 (2.4–3.06) | <0.001 | |
| G/T/A | 0 (0) | 0 (0) | NA | NA | |
| TGF-βa (rs1982037/rs1800471) | C/G | 83 (44.34) | 110 (39.9) | 1.11 (0.75–1.64) | 0.654 |
| T/G | 78 (39.8) | 145 (52.5) | 0.6 (0.4–0.88) | 0.008 | |
| C/C | 27 (13.78) | 21 (7.6) | 1.94 (1.02–3.7) | 0.042 | |
| T/C | 8 (4.08) | 0 (0) | 2.47 (2.21–2.76) | <0.001 | |
NA, not applicable.
Bold values are statistically significant.
C/C and T/C haplotypes in TGF-β (rs1982037, rs1800471) were significantly overrepresented in the patients group comparing to controls while T/G haplotype was significantly less frequent in the patients group.
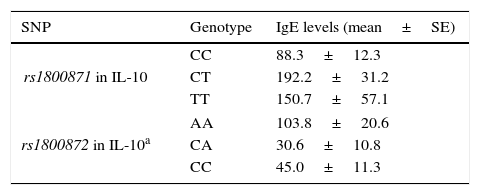
Associations between clinical findings and genotypesThere was not any significant difference in serum IgE levels, eosinophil count and percentage among the genotypic groups of rs1800896, rs1800872 in IL-10 and rs1982037, rs1800471 in TGF-β in the patients with AR. However, patients with CC genotype in rs1800871 in Il-10 had significantly lower levels of IgE (Table 4). In addition, in stratified analysis, AA genotype in rs1800872 was associated with higher IgE levels in patients with mild AR (Table 4).
Significant associations between SNP genotypes and clinical findings in patients with AR.
| SNP | Genotype | IgE levels (mean±SE) |
|---|---|---|
| rs1800871 in IL-10 | CC | 88.3±12.3 |
| CT | 192.2±31.2 | |
| TT | 150.7±57.1 | |
| rs1800872 in IL-10a | AA | 103.8±20.6 |
| CA | 30.6±10.8 | |
| CC | 45.0±11.3 | |
There was not any significant association between different genotypes of the five SNPs and severity or persistency of the disease.
DiscussionAR is an inflammatory disorder of upper airways with a complex interplay of environmental and genetic factors in the background. In this case-control study, we have found a significant relationship between susceptibility to AR and genotypes in three potentially functional SNPs within genes of IL-10 and TGF-β cytokines.
We found that A allele and AA genotype in rs1800896 of IL-10 have a protective effect on AR. A pervious study on Indian allergic patients found that GG genotype in rs1800896 was associated with susceptibility to allergic diseases which was in accordance with the findings of our study.23 In addition, two other studies showed that Egyptian asthmatic children and bronchial asthma patients in Macedonians had significant higher frequency of GG homozygosity genotype in this SNP comparing to healthy individuals.24,25 However, a recent meta-analysis with a total of 4716 asthmatic patients and 5093 controls showed that AA genotype in this SNP was significantly correlated with asthma susceptibility.26 Another systematic review of 11 studies on asthma showed no significant association between this SNP and susceptibility to asthma, but in stratified analysis, G allele had a protective effect on susceptibility to asthma in East Asian populations.27 Inconsistency between the findings of our study and other studies on AR and results of these systematic reviews on asthma might be a result of different roles played by IL-10 in asthma and AR. It has been shown that low levels of IL-10 have a role on the pathogenesis of asthma and airway hyperresponsiveness while a high level of IL-10 has a protective effect against asthma susceptibility.28 Variation in linkage disequilibrium patterns might be another reason for this inconsistency.9
Rs1800896 is located on a region with a putative transcription factor-binding site and, therefore, may influence the binding of transcription factors to the promoter of the IL-10 gene.29 Several in vitro studies reported decreased transcriptional activity in IL-10 promoter in the presence of A allele, although an in vivo study did not support this.29–31 Electrophoretic mobility shift assay (EMSA) studies showed different affinities of alleles of this SNP for a transcription repressor identified as poly ADP-ribose polymerase 1 (PARP-1).30
IL-10 acts mostly as an anti-inflammatory cytokine by its regulatory effects on several pro-inflammatory cytokines. However, it can result in an allergic response by activating B cells and inhibiting IL-12 secretion and T Helper (TH) 1 response.32,33 Therefore, lower levels of IL-10 resulting from A allele in Rs1800896 can have a protective effect on susceptibility to AR.
In our study, frequencies of A/C/A, A/T/C, G/C/A haplotypes were associated with susceptibility to AR while A/T/A and A/C/C haplotypes had fewer frequencies in patients. In vivo studies showed higher levels of IL-10 in individuals with G/C/C haplotype while A/T/A haplotype was associated with the lowest levels of IL-10.30,34
Patients with CC genotype in rs1800871 in IL-10 had lower IgE levels. In addition, AA genotype in rs1800872 was associated with higher IgE levels in patients with mild AR. In accordance with the results of our study, another study on Chinese asthmatic patients found higher levels of IgE in patients with T allele in rs1800871.35 Another study in atopic dermatitis patients showed that the C allele of rs1800872 was associated with decreased total IgE. In vitro studies have shown that IL-10, in the presence of IL4, has a regulatory effect on IgE production and IgE class switching.32,36
TGF-β is a multifunctional cytokine with both pro-inflammatory and anti-inflammatory effects and may modulate the development of allergic inflammation.37 Hemizygosity for the T allele of rs1982037 in TGF-β1 had a protective effect on developing AR, while the C allele and the CG genotype in rs1800471 were associated with a higher susceptibility to AR.
Rs1982037 is a common variant in the first exonic region of TGF-β1 gene which results in substitution of leucine to proline at codon 10 in signal sequence.16 The results of previous studies about the association between alleles in this SNP and serum levels of TGF-β1 were conflicting.38 Rs1800471 is another exonic variant resulting in an Arginine-to-Proline substitution at codon 25.37,38 Arginine in this SNP was associated with higher levels of TGF-β1 in in vivo studies.30
In conclusion, we found that genetic variants in IL-10 and TGF-β are associated with not only the risk of developing AR, but also IgE levels. Considering complex interactions between genetic variants, different genes and environmental factors and different linkage disequilibrium patterns in different ethnic groups, further studies are required to evaluate these associations in different ethnic groups and the effect of these SNPs on cytokine levels in AR patients.
Ethical disclosuresConfidentiality of dataThe authors declare that they have followed the protocols of their work centre on the publication of patient data and that all the patients included in the study have received sufficient information and have given their informed consent in writing to participate in that study.
Right to privacy and informed consentThe authors have obtained the informed consent of the patients and/or subjects mentioned in the article. The author for correspondence is in possession of this document.
Protection of human subjects and animals in researchThe authors declare that the procedures followed were in accordance with the regulations of the responsible Clinical Research Ethics Committee and in accordance with those of the World Medical Association and the Helsinki Declaration.
Conflict of interestThe authors have no conflict of interest to report. The authors did not receive honorarium, grant, or other form of payment to produce the manuscript.