Liver cancer, with high recurrence and metastasis rate, is a common malignant tumor. Circular RNA_0078710 (circ_0078710) has been shown to be take part in the advance of hepatocellular carcinoma. However, the interaction between circ_0091579 and microRNA-431-5p (miR-431-5p) in liver cancer has not been studied.
Materials and methodsThe expressions of circ_0078710, miR-431-5p and Thioredoxin domain-containing 5 (TXNDC5) in liver cancer tissues and cells were detected by quantitative real-time polymerase chain reaction (qRT-PCR). The effect of cric_0078710 in liver cancer cells was assessed by Cell Counting Kit-8 (CCK-8) assay, Transwell, flow cytometry and Dual-luciferase reporter assay. Glycolysis metabolism was examined by lactate production, glucose uptake and ATP level. The protein levels of ki-67, bax and TXNEC5 were tested by western blot. The role of circ_0078710 in vivo was determined by animal study.
ResultsCirc_0078710 and TXNDC5 were notably expressed in liver cancer tissues and cells. Circ_0078710 knockdown diminished proliferation, migration, invasion and glycolytic metabolism of huh7 and Hep3B cells, and accelerated cell apoptosis. MiR-431-5p is the target of circ_0078710, and silence circ_0078710 can inhibit the malignant behavior and glycolysis of hepatocellular carcinoma (HCC) cells by releasing miR-431-5p. In addition, TXNDC5 was a target of miR-431-5p, and overexpression of TXNDC5 restored cell proliferation and glycolysis inhibition due to miR-431-5p. Animal experiments made clear the anti-tumor effect of circ_0078710 knockdown.
ConclusionCirc_0078710 promotes the progression of liver cancer by regulating TXNDC5 expression by targeting miR-431-5p. These results demonstrate that circ_0078710 could be a remedy target for liver cancer.
Liver cancer, with the second highest mortality among all cancers, is one of the malignant tumors, and is characterized by high metastasis and recurrence rates [1–3]. The occurrence of liver cancer is mostly from chronic liver disease transformation, accompanied by liver fibrosis [4]. Hepatocellular carcinoma (HCC), occupying about 90% of all liver cancer patients, is the most common type of liver cancer [2]. However, although liver cancer can be intervened with surgery, drugs or chemotherapy, the prognosis is not good. Therefore, it is very necessary to study the molecular mechanism of liver cancer.
As a competitive endogenous RNA (ceRNA), Circular RNAs (circRNAs) have been actively studied in tumors in recent years [5]. It is a kind of non-coding RNA with a unique continuous covalent closed-loop structure, without terminal 5’cap and 3’tail [6,7]. Several studies have investigated the relationship between circRNAs and liver cancer. For example, Sun et al. showed in vivo and in vitro that silencing circ_0000105 inhibits metastasis and growth of HCC cells [8]. Wang et al. showed that circRHOT1/NR2F6 could be used to regulate the occurrence and development of liver cancer [9]. Xie et al. found that circ_0078710 can intervene in the development of HCC through miR-31 [10]. Nevertheless, the function of circ_0078710 and miR-431-5p in HCC remains unclear.
MicroRNAs (miRNAs), which negatively regulate target genes by interacting with the 3’untranslated region (3’UTR), are also 22-nucleotide non-coding RNAs [11,12]. MiRNAs have been shown to negatively regulate circRNAs, and their interaction can be involved in the growth process of various cancer cells [13,14]. Recently, a large number of literatures have reported the regulatory impact of miRNA-431-5p (miR-431-5p) in the occurrence and progression of certain cancers (such as HCC). For example, miR-431-5p modulates the effects of circ_0001742 in tongue squamous cell carcinoma cells [15]. Studies have shown that miR-431-5p is significantly down-regulated in HCC cells, and its elevation impedes the proliferation of HCC cells [16]. However, the regulatory effect of miR-431-5p and TXNDC5 in liver cancer cells has not been studied.
Thioredoxin domain containing 5 (TXNDC5), a member of the protein disulfide isomerase family, is a fibroblast enriched endoplasmic reticulum (ER) protein [17]. TXNDC5 has been shown to promote the development of many cancers, including HCC. For example, studies have shown that TXNDC5 is significantly upregulated in colorectal cancer tissues [18]. Zang et al. found that circ_0000517/miR-1261-5p/TXNDC5 axis can regulate the progression of liver cancer [19].
In this study, we found that circ_0078710 and TXNDC5 were highly expressed in HCC tissues and cells. In addition, we testify to the role of circ_0078710/miR-431-5p/TXNDC5 axis in liver cancer, in order to reveal the function of circ_0078710 in the progression of liver cancer.
2Methods2.1Patients and cell linesThirty liver tumor specimens and adjacent normal liver tissues were collected from patients with liver cancer in XXXX. The sample was frozen in liquid nitrogen right away. All samples received any treatment preoperatively. Each patient offered written informed consent and received supervision and guidance from XXXX.
Human liver normal cell lines (HHL-5) and human hepatocellular carcinoma cells (HCCLM3, huh7 and Hep3B) were purchased from American Type Culture Collection (ATCC; Manassas, VA, USA), then added 10% (v/v) fetal bovine serum (FBS; GIBCO BRL, Grand Island, NY, USA) and was placed in an incubator containing 5% CO2 at 37°C.
2.2Cell transfectionShort interfering RNA (siRNA) targeting circ_0078710 (si-circ_0078710) and its corresponding control group (si-con), miR-431-5p mimics (miR-431-5p), and its negative control (miR-NC), miR-431-5p inhibitor (anti-miR-431-5p) and its matched (anti-miR-NC), TXNDC5 overexpression vector (pcDNA-TXNDC5) and matched control (pcDNA-NC), siRNA targeting TXNDC5 (si-TXNDC5) and its matched control group (si-NC) were synthesized by Ribobio Co., Ltd. (Guangzhou, China), and then transfected into huh7 and Hep3B cells using Lipofectamine 2000 (Promega, Madison, WI, USA).
2.3Quantitative real-time polymerase chain reaction (qRT-PCR)Trizol Reagent (Thermo Fisher, Waltham, MA, USA) was used to separate total RNA. Then the total RNA was then reverse transcribed using Reverse Transcription Kit (Thermo Fisher). Then, qRT-PCR was performed for cDNA using SYBR Green qRT-PCR Mix (Takara, Shiga, Japan) according to the manufacturer's protocol. GAPDH or U6 were used as internal reference, and the relative expression was calculated by 2−ΔΔCT. The forward and reverse primers were displayed as below: GAPDH, (F: 5’- GGAGCGAGATCCCTCCAAAAT -3’ and R: 5’- GGCTGTTGTCATACTTCTCATGG -3’); U6, (F: 5’- CTCGCTTCGGCAGCACATATACT -3’ and R: 5’- ACGCTTCACGAATTTGCGTGTC -3’); circ_0078710, (F: 5’- CCCGATGACAGGGACAACTG -3’ and R: 5’- GCGGCTTTATATGCCGCTTC -3’); miR-431-5p, (F: 5’- GCCGAGTGTCTTGCAGGCCGT -3’ and R: 5’- CAGTGCGTGTCGTGGAGT -3’); TXNDC5, (F: 5’- TCACTGAGGGAGTACGTGGA -3’ and R: 5’- AGCAGTGCAGTCTACTTCGG -3’).
2.4Western blot analysisThe total protein of liver cancer tissues or cells was separated from using Radioimmunoprecipitation analysis buffer (RIPA; Thermo Fisher). And BCA protein assay kit (Pierce; Rockford, IL, USA) was used to quantify the total protein and loaded into 12% SDS-PAGE. After blocking the membrane with 5% skim milk, anti-β-actin (1:1,000, ab8226, Abcam, Cambridge, MA, USA), anti-TXNDC5 (1:1,000, ab237697, Abcam), anti-Ki-67 (1:2,000, ab15580; Abcam), and anti-bax (1:1,000, ab32503, Abcam) were incubated respectively. Last, Western blot on the membrane was visualized using BeyoECL Plus kit (BeyoTime).
2.5Cell counting Kit-8 assayCCK-8 assayed cell viability. Huh7 and Hep3B were seeded into 96-well plates at a density of 1 × 104 cells per well. Add 10 μL CCK-8 solution (Thermo Fisher) to each well and incubate for 2 hours at 0, 24, 48 and 72 h after inoculation. The absorbance at 450 nm was determined using a microplate reader (BioTek, Winooski, Vermont, USA). Three independent replicates were performed for each CCK-8 test group.
2.6Flow cytometry assayFlow cytometry (BD Biosciences, San Diego, CA, USA) was used to detect the apoptosis of huh7 and Hep3B incubated with Annexin-FITC and Propidium iodide (PI; BD Biosciences) and the cell cycle of huh7 and Hep3B incubated with 1 mL of PI/TritonX-100 staining solution (containing 0.2 mg RNase A, 20 μg of PI, and 0.1% TritonX-100) at 4°C for 30 min.
2.7Transwell assayThe ability of huh7 and Hep3B cells migration and invasion was detected by Transwell. These two difference is that invasion need to be covered with Matrigel in Transwell chambers (Corning, NY, Madison, USA), while migration do not. 5 × 104 cells were added into the upper of Transwell chambers. Meanwhile, the transwell lower chambers was added with fresh culture medium containing 10% FBS. The amount of migrated or invaded cells was counted under a high-powered microscope.
2.8Glycolysis analysesGlycolysis was assessed by monitoring lactate production, glucose uptake and ATP level using commercial kits, including Lactate Assay Kit (Sigma-Aldrich, St. Louis, MO, USA), Glucose Uptake Colorimetric Assay Kit (Sigma-Aldrich) and ATP Bioluminescent Assay Kit (Sigma-Aldrich).
2.9Dual-luciferase reporter assayThe bioinformatics prediction platform circinteractome (https://circinteractome.nia.nih.gov/mirna_target_sites.html) and Starbase (http://starbase.sysu.edu.cn/agoClipRNA.php?source=mRNA) was used to identify the binding sites of between miR-431-5p and circ_0078710 or TXNDC5. The sequences of circ_0078710 or TXNDC5 3’UTR containing presumed miR-431-5p interacting sites were cloned into pGL3-basic vectors (Realgene, Nanjing, China), respectively. Huh7 and Hep3B cells in 48-well plates (8 × 103 cells/well) were transfected with luciferase reporter plasmid in combination with miR-431-5p or control by Lipofectamine 2000 (Promega). The relative luciferase activities were measured by Dual-Luciferase Reporter Assay Kit (GeneCopoeia, Rockville, MD, USA) after transfection 48 h, with Renilla luciferase activity as control.
2.10Immunohistochemical (IHC) staining analysisResected tissues were fixed with 4% buffered paraformaldehyde, dehydrated and embedded in paraffin. Paraffin sections (5 μM) were dewaxed and rehydrated for antigen stripping. Then anti-TXNDC5 was placed at 4°C overnight, and matched with the Goat against mouse IgG (1:10,000, ab205719, Abcam) 1 h. Sections were stained with diaminobenzidine (DAB) kit (Beyotime) according to protocol. The positive staining was observed with a light microscope.
2.11Animal experimentShort hairpin RNA (shRNA) targeting circ_0078710 (sh-circ_0078710) and sh-con were packaged in lentivirus for circ_0078710 stable knockdown by GENESEED. Balb/c mice (female, 6 weeks old) were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China). Sh-circ_0078710 huh7 cells were injected into 1.5 × 106 cells/mice, and the subcutaneous tumor size was measured at week 1, 2, 3, 4 and 5. Tumor volume was calculated according to the formula: volume =1/2 (length × width2). And the mice were sacrificed after measuring the subcutaneous tumor size at fifth week, and the tumor quality was measured. Then the corresponding experiment was carried out.
2.12Statistical analysisGraphPad Prism 7 (GraphPad, San Diego, CA, USA) was utilized for data analysis. Difference comparison between groups were determined using Student's t-test. Pearson's correlation analysis was applied to measure the correlation between two groups. Each experiment was carried out at least three times and the final data were shown as the mean ± standard deviation (SD). Using P<0.05 meant significant difference.
3Results3.1Circ_0078710 and TXNDC5 were highly expressed in liver cancerIn order to investigate the role of circ_0078710 and TXNDC5 in liver cancer, we first detected the expression level of circ_0078710 and TXNDC5 in liver cancer tissues by qRT-PCR. As a result, the expression of circ_0078710 and TXNDC5 were significantly increased in liver cancer tissues (n = 30) (Fig. 1A and B). Similarly, the expression level of TXNDC5 protein was significantly increased in liver cancer tissues (Fig. 1C). Pearson's correlation analysis showed that circ_0078710 was significantly correlated with TXNDC5 and had a positive regulatory relationship (Fig. 1D). In addition, circ_0078710 was analyzed on chr6 with a size of 28899 bp using the cancer-specific circRNA database (Fig. 1E). To sum up, Circ_0078710 and TXNDC5 were up-regulated in liver cancer.
Circ_0078710 and TXNDC5 were up-regulated in liver cancer. (A, B) The expression of circ_0078710 and TXNDC5 in liver cancer tissues (n = 30) were tested by qRT-PCR. (C) Western blot assay detected the expression of TXNDC5 protein. (D) The correlation between circ_0078710 and TXNDC5 expression in liver cancer tissues was analyzed by Pearson's correlation analysis. ***P < 0.001.
In order to further explore the role of circ_0078710 in liver cancer, circ_0078710 was knocked down and transfected into different liver cancer cell lines for detection and analysis. As shown in Fig. 2A, the expression of circ_0078710 was significantly increased in all three different HCC cells. Huh7 and Hep3B cells with the highest expression were selected for subsequent experiments. The transfection efficiency of si-circ_0078710 was detected by qRT-PCR (Fig. 2B). Si-circ_0078710 significantly reduced the cell viability of huh7 and Hep3B cells (Fig. 2C and D). Functionally, silencing circ_0078710 significantly enhanced the apoptosis rate of HCC cells (Fig. 2E). Transwell results showed that knockdown of circ_0078710 inhibited the migration and invasion of HCC cells (Fig. 2F and G). In addition, down-regulation of circ_0078710 significantly reduced glucose uptake, lactic acid production, and ATP levels in huh7 and Hep3B cells (Fig. 2H-J). Western blot results showed that knockdown of circ_0078710 significantly decreased the expression of ki-67 and promoted the up-regulation of bax (Fig. 2K and L). In general, circ_0078710 knockdown can promote apoptosis and inhibit cell viability of HCC cells.
Circ_0078710 knockdown can inhibit the viability, migration and invasion of HCC cells. (A) QRT-PCR was used to detect the expression of circ_0078710 in three different HCC cells. (B) The transfection efficiency of si-circ_0078710 was detected by qRT-PCR. (C, D) CCK-8 was used to detect cell viability. (E) Flow cytometry was used to detect huh7 and Hep3B cells apoptosis. (F, G) Transwell assay was used to assess cell migration and invasion. (H-J) Glucose uptake, lactic acid production, and ATP levels were measured to assess glycolysis metabolism using commercial kits. (K, L) Western blot assay detected the expression of ki-67 and bax protein. ***P < 0.001.
As shown in Fig. 3A, the expression of TXNDC5 was significantly increased in all three different HCC cells. In the same way, huh7 and Hep3B cells with the highest expression were selected for subsequent experiments. The transfection efficiency of si-TXNDC5 was detected by Western blot (Fig. 3B). Si-TXNDC5 significantly reduced the cell viability of huh7 and Hep3B cells (Fig. 3C and D). Functionally, silencing TXNDC5 significantly enhanced the apoptosis rate of HCC cells (Fig. 3E). Transwell results showed that si-TXNDC5 inhibited the migration and invasion of HCC cells (Fig. 3F and G). In addition, si-TXNDC5 significantly reduced glucose uptake, lactic acid production, and ATP levels in huh7 and Hep3B cells (Fig. 3H-J). Western blot results showed that si-TXNDC5 significantly decreased the expression of ki67 and promoted the expression of bax (Fig. 3K and L). In general, si-TXNDC5 can inhibit the viability, migration and invasion of HCC cells.
TXNDC5 knockdown can inhibit the viability, migration and invasion of HCC cells. (A) QRT-PCR was used to detect the expression of TXNDC5 in HCCLM3, huh7 and Hep3B cells. (B) The transfection efficiency of si-TXNDC5 was detected by qRT-PCR. (C, D) CCK-8 was used to detect cell viability. (E) Flow cytometry was used to detect huh7 and Hep3B cells apoptosis. (F, G) Transwell assay was used to assess cell migration and invasion. (H-J) Glucose uptake, lactic acid production, and ATP levels were measured to assess glycolysis metabolism using commercial kits. (K, L) Western blot assay detected the expression of ki67 and bax protein. ***P < 0.001.
We predicted that miR-431-5p was the target of circ_0078710 and that TXNDC5 was the target gene of miR-431-5p using circinteractome (https://circinteractome.nia.nih.gov/mirna_target_sites.html) and starbase (http://starbase.sysu.edu.cn/agoClipRNA.php?source=mRNA) bioinformatics tools, respectively. As shown in Fig. 4A is the binding site of circ_0078710 and miR-431-5p. Dual-luciferase reporter assay results showed that in huh7 and Hep3B cells, the combined transfection of circ_0078710-WT and miR-431-5p significantly inhibited luciferase activity, while there was no significant change in luciferase activity after the combined transfection of circ_0078710-MUT and miR-431-5p (Fig. 4B and C). Next, qRT-PCR was used to detect the co-transfection of si-circ_0078710 and anti-miR-431-5p, and the transfection efficiency was verified by detecting the expression level of miR-431-5p (Fig. 4D and E). The binding sites of miR-431-5p and TXNDC5 are shown in Fig. 4F. Similarly, dual-luciferase reporter assay results showed that in huh7 and Hep3B cells, the combined transfection of TXNDC5 3’UTR-WT and miR-431-5p significantly inhibited luciferase activity, while the combined transfection of TXNDC5 3’UTR-MUT and miR-431-5p showed no significant change in luciferase activity (Fig. 4G and H). Finally, the protein expression of TXNDC5 in huh7 and Hep3B cells after co-transfection of si-circ_0078710 and anti-miR-431-5p was detected by Western blot. The results showed that si-circ_0078710 could significantly reduce the protein level of TXNDC5, but this was relatively recovered with the addition of miR-431-5p inhibitor (Fig. 4I and J). In general, miR-431-5p is the target of circ_0078710, and TXNDC5 is the target gene of miR-431-5p.
MiR-431-5p was target of both circ_0078710 and TXNDC5. (A, F) The binding site of circ_0078710 and miR-431-5p, and the binding sites of miR-431-5p and TXNDC5. (B, C, G, H) Dual-luciferase reporter assays were performed to confirm the association between circ_0078710 and miR-431-5p, and miR-431-5p and TXNDC5 in huh7 and Hep3B cells. (D, E) qRT-PCR was used to detect the expression of miR-431-5p in transfected huh7 and Hep3B cells. (I, J) the protein expression of TXNDC5 in transfected huh7 and Hep3B cells was detected by Western blot. ***P < 0.001.
QRT-PCR was used to detect the transfection efficiency by detecting the expression level of miR-431-5p (Fig. 5A and B). The silencing of miR-431-5p eliminated the inhibitory effect on huh7 and Hep3B cells viability induced by si-circ_0078710 (Fig. 5C and D). Functionally, the addition of miR-431-5p inhibitor can reduce the up-regulated apoptosis rate due to si-circ_0078710 (Fig. 5E and 5F). Meanwhile, the Transwell experiment showed that the addition of anti-miR-431-5p restored the decrease of migration and invasion caused by circ_0078710 knockdown (Fig. 5G and H). Moreover, anti-miR-431-5p significantly up-regulate glucose uptake, lactic acid production, and ATP levels in huh7 and Hep3B cells decreased by si-circ_0078710 (Fig. 5I – K). Functionally, Western blot analysis showed that anti-miR-495-3p could restore the protein levels of ki67 and bax affected by si-circ_0078710 (Fig. 5L and M). In short, miR-431-5p inhibitors can rescue the effect of si-circ_0078710 in HCC cells.
Circ_0078710 regulate the proliferation, migration, and invasion by targeting miR-431-5p in liver cancer cells. (A, B) qRT-PCR was used to detect the expression of miR-431-5p in transfected huh7 and Hep3B cells. (C, D) CCK-8 was used to detect cell viability in transfected huh7 and Hep3B cells. (E, F) Flow cytometry was used to detect huh7 and Hep3B cells apoptosis. (G, H) Transwell assay was used to assess cell migration and cell invasion. (I-K) Glucose uptake, lactic acid production, and ATP levels were measured to assess glycolysis metabolism using commercial kits. (L, M) The protein levels of ki67 and bax were measured by Western blot. ***P < 0.001.
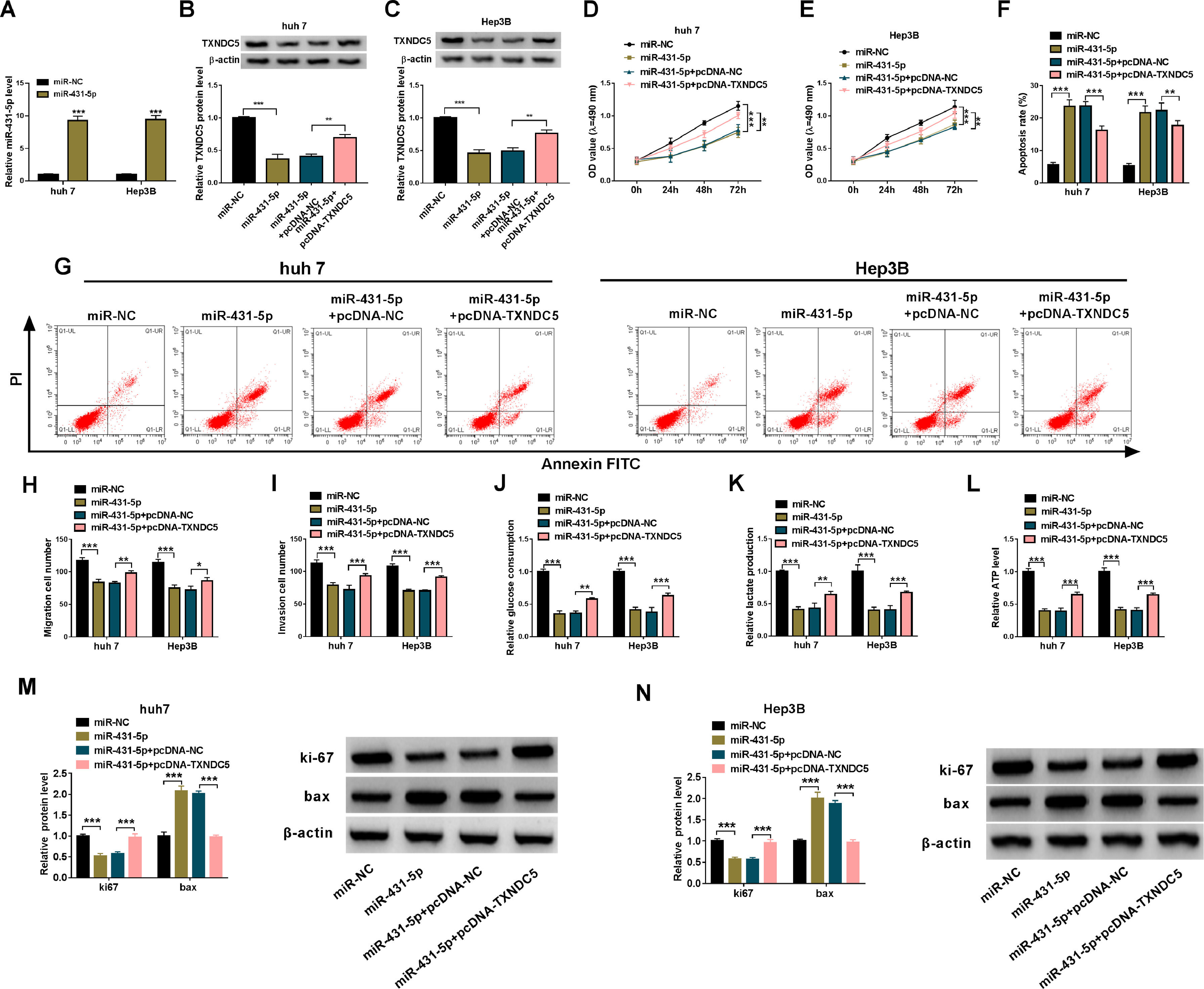
The relationship between miR-431-5p and TXNDC5 was further verified. Firstly, the over-expression efficiency of miR-431-5p was verified by qRT-PCR (Fig. 6A). Next, the expression level of TXNDC5 was detected by Western blot to verify the transfection efficiency of co-transfected miR-431-5p and pcDNA-TXNDC5 (Fig. 6B and C). Functionally, pcDNA-TXNDC5 eliminated the inhibitory effect on huh7 and Hep3B cells viability induced by overexpression of miR-431-5p (Fig. 6D and E). And the transfected huh7 and Hep3B cells apoptosis rate were rescued by pcDNA-TXNDC5 (Fig.6F and G). Meanwhile, the Transwell experiment showed that the addition of pcDNA-TXNDC5 restored the decrease of migration and invasion caused by overexpression of miR-431-5p (Fig. 6H and I). Moreover, pcDNA-TXNDC5 significantly up-regulate glucose uptake, lactic acid production, and ATP levels in huh7 and Hep3B cells decreased by overexpression of miR-431-5p (Fig. 6J – L). Functionally, Western blot analysis showed that pcDNA-TXNDC5 could restore the protein levels of ki67 and bax affected by overexpression of miR-431-5p (Fig. 6M and N). In short, pcDNA-TXNDC5 can rescue the effect of overexpression of miR-431-5p in HCC cells.
TXNDC5 regulate the proliferation, migration, and invasion by targeting miR-431-5p in liver cancer cells. (A) QRT-PCR was used to detect the expression of miR-431-5p in transfected huh7 and Hep3B cells. (B, C) Western blot was used to detect the expression of TXNDC5 in transfected huh7 and Hep3B cells. (D, E) CCK-8 was used to detect cell viability in transfected huh7 and Hep3B cells. (F, G) Flow cytometry was used to detect huh7 and Hep3B cells apoptosis. (H, I) Transwell assay was used to assess cell migration and cell invasion. (J-L) Glucose uptake, lactic acid production, and ATP levels were measured to assess glycolysis metabolism using commercial kits. (M, N) The protein levels of ki67 and bax were measured by Western blot. ***P < 0.001.
In order to better explore the effect of circ_0078710 on liver cancer, we constructed a xenotransplantation model to explore the effect of circ_0078710 knockdown in liver cancer tumor. By recording and observing tumor volume (Fig. 7A) and volume weight (Fig. 7B), we found that huh7 cells transfected with sh-circ_0078710 could inhibit tumor growth in a transplanted tumor model. Then, the expression of circ_0078710 and miR-431-5p was analyzed by qRT-PCR, and the results showed that knockdown of circ_0078710 could significantly decrease the expression of circ_0078710 and enhance the expression of miR-431-5p (Fig. 7C and 7D). And Western blot analysis also confirmed that sh-circ_0078710 remarkably reduced the TXNDC5 protein level (Fig. 7E). Lastly, the immunoreactive levels of TXNDC5 were downregulated in tumor tissues from sh-circ_0078710 group compared with sh-con group (Fig. 7F). In short, circ_0078710 knockdown reduced the growth of liver cancer tumor.
Circ_0078710 knockdown inhibited tumor growth in vivo. (A, B) Tumor volume and weight after circ_0078710 knockdown in vivo. (C, D) Relative expression levels of circ_0078710 and miR-431-5p in xenografts were detected by qRT-PCR. (E) TXNDC5 protein level tested by western blot in xenografts. (F) The levels of TXNDC5 was evaluated by immunohistochemical staining in tumor samples. ***P < 0.001.
Liver cancer is a malignant tumor disease that seriously threatens human health, with high mortality rate and difficult treatment. More and more reports have shown that circRNAs, as a newly discovered endogenous non-coding RNA, play an indispensable role numerous cancers, such as breast cancer [20], ovarian cancer [21], esophageal cancer [22] and liver cancer [9]. However, there are relatively few studies on circ_0078710 in cancer. For instance, circ_0078710 was highly expressed in HCC patients, witch consistent with our study [10]. And Xie et al. found that abnormal up-regulation of circ_0078710 in HCC cell lines promoted malignant behavior and tumor growth [10]. Similarly, we found that cell proliferation, migration and invasion ability of huh7 and Hep3B cells could be significantly inhibited and the apoptotic ability of cells was promoted by silencing circ_0078710. In addition, we also found that down-regulation of circ_0078710 inhibited glycolytic metabolism. Animal experiments showed that knocking down circ_0078710 could significantly inhibit tumor growth in nude mice in vivo. These results suggested that circ_0078710 could be a potential therapeutic target for hepatocellular carcinoma.
Our mechanistic analysis showed that miR-431-5p was a target of circ_0078710. Previous studies found that the recovery of miR-431-5p eliminated the cancer-promoting effect of ATG3 in colon cancer [23]. In addition, miR-431-5p was significantly down-regulated in both HCC cells and tissues, and overexpression of miR-431-5p significantly inhibited the activity and metastasis of HCC cells by inhibiting UROC28 gene [16]. Consistently with the above study results, we found that inhibition of miR-431-5p recovered the prohibitive effects of silencing circ_0078710 on proliferation, migration, invasion and glycolytic metabolism of HCC cells. These showed that miR-431-5p may interact with circ_0078710 to play a role in liver cancer.
It is well known that TXNDC5 is an oncogene that is up-regulated in many malignant tumors, such as HCC, castration-resistant prostate cancer, and esophageal squamous cell carcinoma [19,24-26]. In this study, we demonstrated that TXNDC5 is a target of miR-431-5p. Similarly, Zang and colleagues found that miR-1296-5p plays a tumor-promoting role in HCC by targeting TXNDC5, and TXNDC5 is highly expressed in HCC, which is consistent with our results [19]. Similarly, Yu et al. found that TXNDC5 was highly expressed in HCC, and that TXNDC5 overexpression played a catalytic role in malignant behavior of HCC cells [24]. Our results also support this conclusion, TXNDC5 unusual increased restored the inhibition of miR-431-5p on the proliferation, migration, invasion and glycolytic metabolism of huh7 and Hep3B cells. More importantly, circ_0078710 knockdown significantly weakened TXNDC5 expression by releasing miR-431-5p, suggesting that circ_0078710 regulates the miR-431-5p/TXNDC5 pathway.
In summary, we found that the expression of circ_0078710 was up-regulated in liver cancer. Circ_0078710 knockdown inhibited the malignant behavior of HCC cells. We demonstrated that down-regulation of circ_0078710 partially inhibits the progression of liver cancer by regulating the miR-431-5p/TXNDC5 pathway. This study suggests that inhibition of circ_0078710 may be a promising strategy for the treatment of HCC.
FundingNone.
None.