Background & aims. G-allele of PNPLA3 (rs738409) favours triglycerides accumulation and steatosis. In this study, we examined the effect of quercetin and natural extracts from mushroom and artichoke on reducing lipid accumulation in hepatic cells.
Material and methods. Huh7.5 cells were exposed to oleic acid (OA) and treated with quercetin and extracts to observe the lipid accumulation, the intracellular-TG concentration and the LD size. Sterol regulatory element binding proteins-1 (SREBP-1), peroxisome proliferator-activated receptor (PPARα-γ) and cholesterol acyltransferase (ACAT) gene expression levels were analysed.
Results. Quercetin decreased the intracellular lipids, LD size and the levels of intracellular-TG through the down-regulation of SREBP-1c, PPARγand ACAT1 increasing PPARα. The natural-extracts suppressed OA-induced lipid accumulation and the intracellular-TG. They down-regulate the hepatic lipogenesis through SREBP-1c, besides the activation of lipolysis through the increasing of PPARα expression.
Conclusions. Quercetin and the aqueous extracts decrease intracellular lipid accumulation by down-regulation of lipogenesis and up-regulation of lipolysis.
Non-alcoholic fatty liver disease (NAFLD) is defined as the accumulation of excessive fat in the liver in the absence of excessive drinking of alcohol.1 It has been included as a metabolic abnormality such as obesity, type 2 diabetes, arterial hypertension, and hypertriglyceridemia.2,3 NAFLD encompasses a wide spectrum of liver damage ranging from simple steatosis to non-alcoholic steatohepatitis (NASH), advanced fibrosis, and rarely, progression to cirrhosis and hepatocellular carcinoma.4
Hepatic steatosis can either be a benign, non-inflammatory condition, or can be associated with non-alcoholic steatohepatitis (NASH). The earliest stage is characterized by the excessive triglycerides (TGs) accumulation as lipid droplets (LDs) in the cytoplasm of hepatocytes. Hepatic steatosis is often self-limited, however it can progress to NASH, which is known by the presence of hepatocyte injury (hepatocyte ballooning and cell death), inflammatory infiltrate, and/or collagen deposition (fibrosis).5 It has been postulated that the inhibition of excessive lipid synthesis and uptake could be an effective intervention for NASH.6,7
Dietary fat is one of the most important environmental factors associated with the incidence of NAFLD. The search of functional food ingredients such as herbal extracts or flavonoids capable to suppress the accumulation of hepatic lipid8–12 by the modulation of several pathways is ongoing. Several polyphenols and phenolic compounds, such anthocyanins, curcumin, resveratrol, silymarin and those present in coffee and tea have been proposed as NAFLD treatment but the varying bioavailability remains poor, so further studies are needed for the future clinical applications.13
Steatosis could be modulated by genetic susceptibility.14 In 2008, Romeo, et al. performed an independent genome-wide association study to identify genetic determinants of liver steatosis.15 The author figured out that the polymorphism rs738409 C>G in PNPLA3 gene was robustly associated with an increased risk for hepatic steatosis. Patatin-like phospholipase domain-containing protein 3 (PNPLA3) is a transmembrane protein expressed prominently in hepatocytes and the substitution I148M has been suggested to impair TG hydrolysis in hepatocytes, favouring its accumulation.16 Recently it has been proposed that PNPLA3-148M evade ubiquitylation and proteasomal degradation, resulting in the accumulation of PNPLA3-148M on the surfaces of lipid droplets impairing TGs mobilization from LDs.17 The discovery of new drugs to reduce the risk of NAFLD would be useful considering the genetics factors. The purposes of this study were to elucidate the role of quercetin and other water-soluble extracts in an in-vitro model with unfavourable genotype GG for PNPLA3.18
Material and MethodsCell CultureHuh7.5 cells were routinely cultured in DMEM (ThermoFisher, MA, USA) supplemented with 10% FBS and 1% penicillin-streptomycin in an incubator under an atmosphere of 5% CO2 at 37 °C. The Huh7.5 cell model of OA-induced intracellular lipid accumulation was developed as previously described.19 Cells were cultured with 1mM of Oleic acid for 48 h. Control cells were treated with FFA-free medium containing the vehicle dimethyl sulfoxide (DMSO).
PNPLA3 genotypingDNA from cells was extracted using the DNA isolation Kit with the MagNA Pure LC Instruments (Roche). The rs738409 SNPs were analysed using the StepOnePlus Real Time PCR System (Applied Biosystem, Foster City, USA) with a Taqman SNP Genotyping Assay, using published sequences from the NCBI Entrez SNP Database (www.ncbi.nlm.nih.gov/sites/entrez) (rs738409: 5’-AAG-GAGGGATAAGGCCACTGTA-3 as forward and 5’-CTTTCACAGGCCTTGGTATGTTC-3’ as reverse primer).
Preparation of aqueous extractsWhite button mushrooms (Agaricus bisporus) were used as raw material after cultivation in a pilot plant at the University of Seville (Spain), according to standard procedures. All chemicals used were of analytical grade. Agaricus bisporus Aqueous Extract (AbAE) was obtained by an enzymatic procedure based on the protocol described by Cremades and colleagues.20 Briefly, after A. bisporus homogenization (10g + 10 mL distilled water) and enzymatic digestion with a mixture of glucanase and chitinase enzymes (Novo Nordisk®, Denmark) at pH = 5, temperature 55 °C and an enzyme/substrate ratio of 0.01, for 24 h. Finally, temperature was raised up to 90 °C for 120 min to inactivate the enzymes. After cooling to room temperature, pH was adjusted to 7.0 with 1M NaOH and centrifuged at 8000 × g. The supernatant was collected and filtered through a 0.2 µm membrane, using the filtrate as “crude AbAE” for activity assays. Cynara scolymus aqueous extracts were obtained by a similar procedure but using a mixture of celluloses and proteases as hydrolytic agent.20
Detection of LDs by Fluorescent Microscopy Oil Red O (ORO) cell staining.Neutral lipids stored into the LDs were visualized by fluorescence microscopy using ORO staining (SigmaAldrich, MO, USA), a vital lipophilic dye used to label fat accumulation in the cytosol, according to Hu and colleagues.21 To analyze fat accumulation 20,000 Huh7.5 cells were seeded and grown on coverslips in 24 wells plate. Huh7.5 cells were cultured in the presence of oleic acid 1mM diluted in DMSO (less than 0.01% of total volume) and treated for 48h with 50 µM of quercetin (HWI ANALYTIK, GmbH, Germany) or 0.1 mg/mL of water-soluble extract from A. Bisporus (M) or Cynara scolymus (A). The cells were rinsed two times with phosphate-buffered saline (PBS) pH 7.4, fixed with 4% paraformaldehyde in PBS for 10min and permeabilised with 0.2% Triton X-100 for 2 min. Nuclei cells were stained with 46-diamidino- 2phenylindole (DAPI) for 30min at room temperature, and neutral lipids were stained with ORO as previously described.22 Images were acquired with a fluorescence microscope (OLYMPUS BX41) equipped with the standard epifluorescence filter set up for DAPI and FITC. For determination of LD diameter images were captured under oil with a 100x plan apochromat objective. Analyses were performed on three independent experiments measuring at least 100 cells for each treatment using Imaging Software cell^F (Olympus, Tokyo, Japan).
Fluorimetric determination of intracellular fat content - Nile red stainingThe intracellular fat content was determined fluorimetrically based on Nile Red staining, a vital lipophilic dye used to label fat accumulation in the cytosol.23 Ten thousand Huh7.5 cells were grown in 96 plate wells, exposed to oleic acid and treated with quercetin and water-soluble M or A for 48 h. AdipoRed™ Reagent (Lonza, Basel, Switzerland) was added to each well and incubated at room temperature for 10 min. Intensity fluorescence was quantified using Synergy HT at 485/572 nm (BioTeK, VT, USA) and normalized to protein concentration.
RNA isolation, retro-transcription and quantitative polymerase chain reaction (q-PCR)Total RNA was extracted from the above-treated cells using the guanidine isothiocyanate method.24 RNA samples were treated with DNaseI. Total RNA was subjected to reverse transcription (RT) using commercially available kits (QuantiTect Rev. Transcription Kit; Qiagen, Hilden, Germany) according to the manufacturer’s instructions. SREBP-lc, PPARγ, PPARα, ACAT, DGAT-1, DGAT-2, FASN, MTTP, APOB and APOE gene expression levels were analysed using commercial oligonucleotides (Qiagen, Hilden, Germany).
Statistical AnalysisContinuous variables are described as means ± SD of minimum three independent experiments. The Student t-test was used for comparisons between groups. P values P < 0.05 (*) p < 0.01 (**) and p < 0.001 (***) were considered statistically significant.
ResultsHuh7.5 PNPLA3 genotypeHuh7.5 cells presented unfavourable genotype GG for PNPLA3 as previously was showed.18
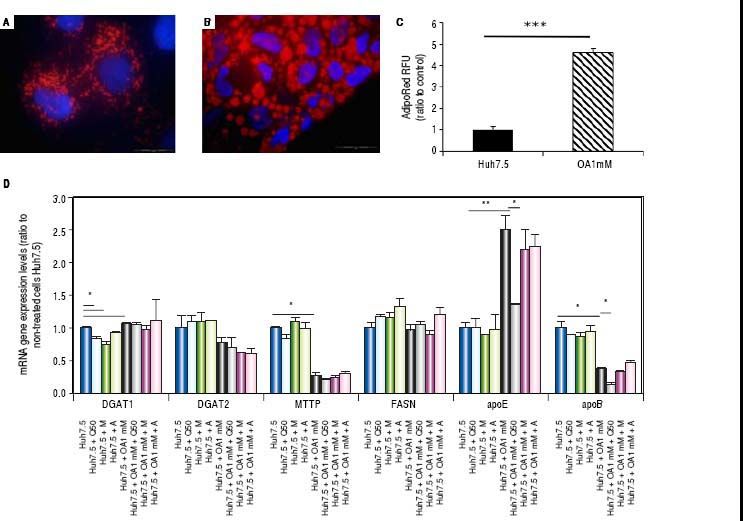
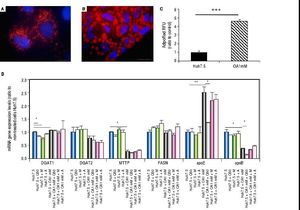
Oleic acid-induced intracellular lipid accumulation in Huh7.5 cellsUsing oil red staining we observed that untreated Huh7.5 cells revealed almost absence of intracellular lipid (Figure 1A). However, as shown in figure 1B, after OA exposure LD were higher in number and size.
Untreated Huh7.5 (A) and Huh7.5 cells cultured at OA concentrations 1 mM, for 48 hours (B). LDs and nuclei were stained with ORO (red) and Dapi (blue), respectively. Images were acquired with a fluorescence microscope (OLIMPUS BX41) equipped with the standard epifluorescence filter set up for DAPI and FITC under oil with a 100x plan apochromat objective. C. Intracellular triglycerides concentration. AdipoRed™ Assay of Huh7.5 and OA-Huh7.5 cells. The intracellular triglycerides were stained and the concentration of triglycerides was quantified by fluorescence. (Huh7.5 fold=1) (***p < 0.01). Data are presented as the mean values ± SD obtained from three independent experiments. D. (A) mRNA gene expression levels of DGAT-1, DGAT-2, MTTP, FASN, apoE and apoB were determined by RT-PCR. Results were normalized using GAPDH and Huh7.5 non-treated cells were used as reference. *p < 0.05; **p < 0.01 and ***p < 0.001. Data are the mean value ± SD obtained from three independent experiments.
Using a fluorescence-based AdipoRedTM assay, the amount of intracellular lipids increased in the presence of OA 78.31 ± 3.78% compared to untreated cells (Figure 1С).
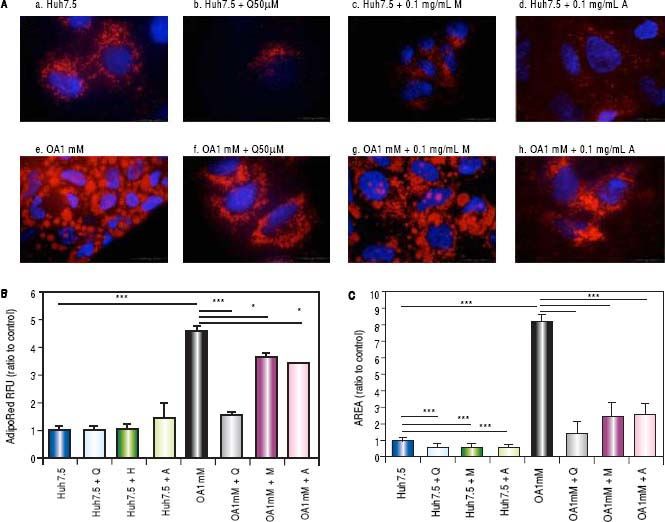
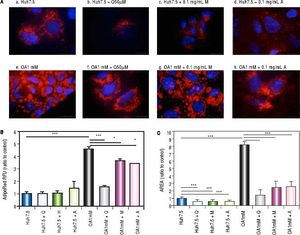
Effects of Quercetin and water-soluble extracts from mushroom and artichoke on hepatic lipid accumulationFirst, we have determined the dose by cytotoxic assay and at 0.1 mg/mL no toxic effects were observed (data not shown). A significant reduction in lipid accumulation was observed by microscopy after quercetin or extracts addition (Figure 2A). After treatment, a significant reduction of the LD size in OA-treated cells was found (Figure 2B). Intracellular lipid concentration was decreased in OA-treated cells by quercetin (66 ± 2.04%), M aqueous extract (20.40 ± 3.63%) and A aqueous extract (24.61± 0.19%) (Figure 2C).
A. Huh7.5 (a) and OA-Huh7.5 (e) cells treated with quercetin 50 µМ (b-f), aqueous extracts from mushroom (M) (c-g) and artichoke (A) (d-h) (0.1 mg/mL), for 48 h. LDs and nuclei were stained with ORO (red) and Dapi (blue), respectively. Images were taken using a fluorescence microscope (OLIMPUS BX41) equipped with a 100x objective and the standard epifluorescence filter set up for DAPI and FITC. B. LDs size were measured by Imaging Software cell^F Software. Results are expressed as fold-area µm3. C. Intracellular triglycerides concentration. AdipoRed Assay. The intracellular triglycerides were stained and the concentration of triglycerides was quantifed by fluorescence (Huh7.5 fold = 1). Data are presented as the mean values ± SD obtained from three independent experiments. Experiments were performed in triplicate. (*) p < 0.05; (**) p < 0.01 (***) p < 0.001.
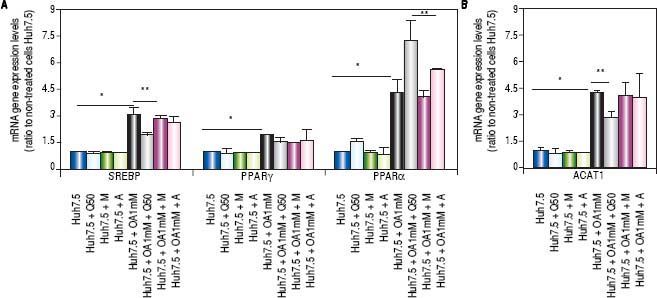
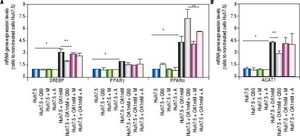
Genes involved in lipogenesis (SREBP-1c, PPARγ and PPARα), significantly increased in OA-induced Huh7.5 cells at the mRNA levels (p < 0.05) (Figure 3). As shown in figure 3A, quercetin treatment decreased the expression of SREBP-1c (fold inhibition: 1.57 ± 0.13) and PPARy (fold inhibition: 0.77 ± 0.10). The same effect was observed after treatment with the aqueous-extracts. Conversely, quercetin and artichoke extract increased the gene expression of PPARα (Figure 3A). ACAT gene expression was increased by OA 1mM and decreased significantly (fold inhibition: 1.48 ± 0.3) after quercetin treatment being this reduction less significantly after extract addition (Figure 3B). Genes implicated on triglycerides and VLDL pathways were also modulated in OA-induced Huh7.5 cells. DGAT-l and APOE mRNA levels were significantly increased and MTTP and APOB were decreased in OA-induced Huh7.5 cells (p > 0.05). Quercetin and artichoke extract decreased DGAT1 mRNA levels in Huh7.5 and the apolipoproteins genes were repressed by quercetin in the OA-induced model (Figure 1D).
A.mRNA gene expression levels of SREBP-1c, PPARγand PPARαwere determined by RT-PCR. B. ACAT-1 mRNA gene expresision levels. Results were normalized using GAPDH and Huh7.5 non-treated cells were used as reference. *p < 0.05; **p < 0.01 and ***p < 0.001. Data are the mean value ± SD obtained from three independent experiments.
Current NAFLD therapies include lifestyle modifications, physical activity and medical intervention. Probiotics, functional food and several natural compounds (i.e. resveratrol and quercetin,25 anthocyanins,26 vitamin E27) may be promising in helping therapeutic approaches. On the basis of these data, it seems that foods rich in quercetin and/or including natural extracts from mushroom and artichoke may be useful for the prevention of NAFLD.
In addition to resveratrol and quercetin, other polyphenols such as Anthocyanin Cy-3-g, Proanthocyanidins, Theaflavin (a flavan-3-ol) and Ellagic acid have been studied as potential agent for both prevention and treatment of hepatic steatosis.28–31 Resveratrol is a stilbene occurring naturally in several plants and provided in the diet by various foodstuffs. In recent years, it has been shown to modify lipid metabolism, and more specifically to induce a reduction in liver triacylglycerol content.32–34
Quercetin is a natural polyphenol belonging to a group with a variable structure, known as flavonoids. It is found in onions, broccoli, tomatoes, apples and berries. Several studies have shown that quercetin has more than one effect in order to modify the intracellular lipid content.11 Vidyashankar and colleagues35 showed that quercetin decreased triacylglycerol accumulation, modulated the insulin resistance, inflammatory cytokine secretion, and increased cellular antioxidants suggesting that quercetin could be an effective molecule for NAFLD.
In this study, we have shown that aqueous extracts from mushroom and artichoke may be useful for therapeutic interventions in lipid accumulation-related liver pathologies like NAFLD. Beneficial effects of polyphenols in the prevention and treatment of liver steatosis have been widely reported.25,36 These molecules present hepaticprotective effects because they reduce liver fat accumulation, mainly by lowering lipogenesis and increasing fatty acid oxidation. Besides, it has been shown that polyphenols are able to reduce oxidative stress and inflammation, the main factors responsible for liver damage.37 To date, these beneficial effects have been demonstrated in cultured cells and animal models.
Quercetin was used as a reference or like a positive control.11 Our results confirm that quercetin reduces TGs concentration and LDs size in an OA in vitro model. The use of aqueous extracts showed similar effects than quercetin such as an important reduction of intracellular lipid content, LD size and intracellular triglycerides concentration.
Lipid accumulation in the liver may be caused by enhanced de novo lipogenesis, activation of lipid uptake, and lowering of lipid catabolism. Fatty acids are known to be ligands for nuclear transcription factors, such as SREBP-1c, PPARγ and PPARα.38 It has been reported that PPARα knockout (-/-) mice developed severe hepatic steatosis upon fasting as a result of failure to up-regulate the fatty acid oxidation pathway.39PPARα activation is required to enhance hepatic lipid turnover to enable sufficient clearance of lipids from the liver, preventing lipid accumulation and peroxidation in murine NASH models.40 Our result confirmed that the therapeutic effect of quercetin on lipid metabolism in Huh7.5-induced fatty liver cells is partly due to PPARα upregulation (inducing lipolysis) and SREBP-1c downregulation (reducing lipogenesis).41,42Cynara scolymus extract increased PPARα gene expression levels in the model of steatosis which controls fatty acid degradation. Anderson, et al. showed that pathogenesis of NASH increased the pool of free fatty acids through de novo lipid synthesis and nuclear receptors activation (SREBP-1, ChREBP-1, and PPARγ).43 In our study, OA significantly increased SREPB-lc gene expression which was disrupted by quercetin. In addition, PPARγ gene expression was induced by OA and this effect was diminished after treatment. Beside, we demonstrated that OA modified genes involved in TGs synthesis and VLDL secretion pathway however quercetin and aqueous extracts treatment did not affect on their expression. This work demonstrated that Agaricus bisporus and Cynara scolymus aqueous extracts, together with quercetin treatment, modified nuclear transcription factors leading to a significant decrease of intracellular lipid content and LDs size.
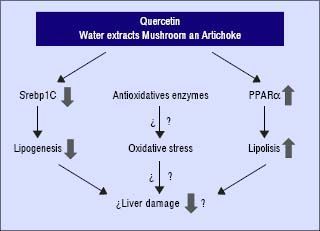
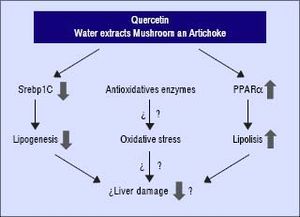
In conclusion, these compounds may interfere and prevent the development of NAFLD in the presence of unfavourable genotype GG of PNPLA3. Quercetin and the aqueous extracts (A and M) may prevent the progression of liver damage related to NAFLD by two independent mechanisms: inhibition of lipogenesis by reducing SREBP-lc and promoting lipolysis through PPARα induction (Figure 4). Further studies are required to clarify the molecular mechanism including their role in the oxidative stress. In addition, it would be needed to test the specific effect of single compounds which are included in the aqueous extracts described in this work.
Abbreviations- •
A: Cynara scolymus.
- •
AbAE: Agaricus bisporus aqueous extract.
- •
ACAT: Cholesterol acyltransferase.
- •
APOB: Apolipoprotein-B.
- •
APOE: Apolipoprotein-E.
- •
DAPI: 46-diamidino- 2-phenylindole.
- •
DGAT-1: Diacylglycerol acyltransferase-1.
- •
DGAT-2: Diacylglycerol acyltransferase-2.
- •
DMEM: Dulbecco’s Modified Eagle’s Medium.
- •
DMSO: Dimethyl sulfoxide.
- •
FASN: Fatty Acid Synthase.
- •
FBS: Fetal bovine serum.
- •
FFA: Free fatty acid.
- •
FITC: Fluorescein isothiocyanate.
- •
LDs: Lipid droplets.
- •
M: Agaricus Bisporus.
- •
MTTP: Microsomal triglyceride transfer protein.
- •
NAFLD: Non-alcoholic fatty liver disease,
- •
NASH: Non-alcoholic steatohepatitis.
- •
OA: Oleic acid.
- •
ORO: Oil Red O.
- •
PBS: Phosphate-buffered saline.
- •
PNPLA3: Patatin-like phospholipase domain-containing protein 3.
- •
PPAR-a: Peroxisome proliferator-activated receptor alpha.
- •
PPAR-?: Peroxisome proliferator-activated receptor gamma.
- •
q-PCR: Quantitative polymerase chain reaction.
- •
RT: Retro-transcription.
- •
SD: Standard deviation.
- •
SREBP: Sterol regulatory element binding protein-1.
- •
TG: Triglycerides.
- •
Financial support. None.
None.
Specific Author Contribution- •
Planning and conducting the study: Ángela Rojas, Manuel Romero-Gómez.
- •
Drafting the manuscript: Ángela Rojas, Jose Antonio del Campo, Manuel Romero-Gomez.
- •
Interpreting data: Jose Antonio del Campo, Juan Bautista, Manuel Romero-Gomez.
- •
Performing in vitro studies: Ángela Rojas, Paloma Gallego, Antonio Gil-Gómez, Rocío Muñoz, Lourdes Rojas, Rosario Maldonado, Rocío Gallego-Durán, Marta García-Valdecasas.