Background and aims. CD4+ T cells play an important role in response to hepatitis B virus (HBV) infection. We investigated the change in CD4+ T-cell subpopulations and viral load in patients with chronic HBV infection who were treated with entecavir.
Material and methods. Thirty patients with chronic HBV infection were enrolled according to the criteria recommended by the Chinese Society of Infectious Diseases and the Chinese Society of Hepatology. The expressions of signature transcription factors and cytokines of CD4+ T-cell subpopulations were measured in chronic hepatitis B (CHB) patients treated with entecavir treatment.
Results. Entecavir treatment significantly attenuated hepatitis B virus DNA load and affected the CD4+ T-cell subsets in CHB patients. A dramatic decrease in the Th17 and Treg cell frequencies and expressions of their related cytokines were found in CHB patients with entecavir treatment. In contrast, entecavir treatment caused a remarkable increase in the Th2 cell frequencies and expressions of their related cytokines.
Conclusion. Our results suggested that Th17 and Treg cells were the more sensitive subtypes to entecavir-induced inhibition of HBV replication compared to Th1 and Th2 cells in chronic HBV patients.
Hepatitis B virus (HBV) causes the disease hepatitis B.1 An estimated two billion people worldwide have been infected with HBV, and approximately 5% of the world’s populations (350 million people) are chronic HBV patients.2 Chronic hepatitis B (CHB) virus is a common cause of death associated with liver failure, cirrhosis and hepatocellular carcinoma (HCC).3 An estimated 620,000 people die annually from hepatitis B-associated acute and chronic liver diseases.4
Both the innate and adaptive immune responses are known to be involved in viral clearance during HBV infection.5 There is a clear dichotomy in the profile of the immune responses observed, depending on whether patients naturally resolve viral infection or develop chronic infection. Patients suffering from chronic infection have very weak or functionally impaired immune responses. This condition of T cell hyporesponsiveness is frequently associated with high viral and/or antigen load in chronic hepatitis B patients.6,7 If we understand the characteristics of cell-mediated immune responses and the immunosuppressive environment generated during chronic HBV infection, we will describe current and future approaches for manipulation of the immune system to achieve sustained control of this persistent infection.
Approved available drugs for HBV infection include interferon alpha-α, pegylated interferon alpha-α (Pegasys®), lamivudine (Epivir®), adefovir (Hepsera®), telbivudine (Tyzeka®), tonofovir (Viread®) and entecavir (Baraclude®). In addition, there are several available vaccines against HBV infection. Studies have shown that entecavir can be used to treat chronic hepatitis B and human immunodeficiency virus (HIV) infection.8–10 Entecavir can inhibit HBV replication by interfering with HBV DNA polymerase, an enzyme needed by the virus to replicate in liver cells.8 Entecavir may help to lower the amount of HBV and can reduce the ability of the HBV to multiply and infect new liver cells.
Patients with chronic infection were significantly reduced in HBV-specific CD4+ and CD8+ T cell responses, compared with the infections of those in individuals who successfully overcame HBV infection. Generally, patients with a chronic HBV infection exhibit a weak or undetectable HBV-specific T cell response.11 An increase in HBV-specific CD4+ and CD8+ T-cell responses is known during the treatment of chronic HBV infection. The CD8+ T-cell response is known to be important cellular effectors to modulate HBV infection by the present of CD4+ T-cells serving as the master regulators of the adaptive immune response.12 Therefore, HBV-specific CD4+ T cell response plays an important role in the anti-HBV infection. Many studies have shown that entecavir can affect the dynamics of CD4+ T cell reconstitution and changes in immune activation in patients with HBV infection.13–15 However, there is little evidence as to whether entecavir can affect the dynamics of CD4+ T cell reconstitution and changes in immune activation in patients with chronic HBV infection. Furthermore, at least four distinct CD4+ T-cell subsets have been shown to exist (Th1, Th2, Treg, and Th17 cells). Dissecting the precise roles of each CD4+ T-cell subset will improve our understanding of hepatitis pathogenesis. In order to understand the precise roles of each CD4+ T-cell subset in patients with chronic HBV infection, we investigated the change in CD4+ T-cell subpopulations and viral load in patients with chronic HBV infection who were treated with entecavir. In this study, we report that Th17 and Treg cells were the most sensitive subpopulations of the peripheral blood and liver tissue CD4+ T cells to entecavir treatment over 52 weeks.
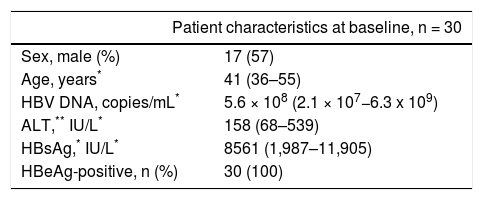
Material and MethodsPatients and healthy volunteersThirty patients with chronic HBV infection were enrolled from the Department of Infectious Diseases, Southwest Hospital, Chongqing, China. CHB was diagnosed according to the criteria recommended by the Chinese Society of Infectious Diseases and the Chinese Society of Hepatology.16 The clinic pathological data about the CHB patients are shown in table 1. Heparinized blood samples and liver biopsies were collected from patients at baseline 0, 12, 24, 36, and 52 weeks after entecavir treatment (0.5 mg orally per day). Plasma samples from healthy volunteers (17 males and 13 females, aged 36-55 years) were used to determine the normal values of the indicators. Plasma HBsAg concentration was detected by ELISA. Plasma ALT and AST was measured by the CHEMIX-180 automated biochemistry analyzer (Japanese SYSMEX Corporation, Japan). The protocol was approved by the Ethical Committee of Southwest Hospital. Written informed consent was obtained from each patient before entering the study, according to protocol.
Clinical characteristic of the CHB patients at study enrollment.
| Patient characteristics at baseline, n = 30 | |
|---|---|
| Sex, male (%) | 17 (57) |
| Age, years* | 41 (36–55) |
| HBV DNA, copies/mL* | 5.6 × 108 (2.1 × 107−6.3 x 109) |
| ALT,** IU/L* | 158 (68–539) |
| HBsAg,* IU/L* | 8561 (1,987–11,905) |
| HBeAg-positive, n (%) | 30 (100) |
ALT: alanine aminotransferase. AST: aspartate aminotransferase. HBsAg: hepatitis B surface antigen.
In order to separate fresh heparinized peripheral blood from the peripheral blood mononuclear cells (PBMCs), we used the Optipep Nycoprep Lymphoprep kit (Axis-Shield, Oslo, Norway) according to the manufacturer’s protocol. CD4+ T cells were sorted from PBMCs using CD4+ T Cell Isolation Kit (Miltenyi biotec, Germany) according to the manufacturer’s protocol. Total RNA was isolated from CD4+ T cells using TRIzol reagent (Lift Technologies, USA) according to the manufacturer’s instructions, and frozen in −70 °C until used for mRNA extraction. The plasma from the same peripheral blood sample was collected and frozen in −70 °C until used for the enzyme-linked immunosorbent assay (ELISA).
Quantitative real-time RT-PCRIsolated RNA was subjected to reverse transcription following the manufacturer’s protocol from Toyobo (Osaka, Japan). Real-time PCR primers for human FOXP3, T-bet, RORγt, GATA3, and GAPDH were designed and synthesized by Sangon Biotech Co., Ltd (Shanghai, China) (Table 2). The real-time PCR reactions were processed using SensiMix two-step RT-PCR with SYBR Green (Quantace, London, UK), according to the manufacturer’s protocol. Each mRNA expression level was normalized to the GAPDH mRNA expression in the respective cDNA preparation. Controls were arbitrarily set to 1.0, and gene induction was calculated according to the degree of fold increase compared to the controls. The quantitation of HBV DNA in patients serum was performed by PCR based on Kaneko’s method.17 The method measuring range is 3 × 102 −3 × 1012 copies/mL.
Sequence of primers used for quantitative RT-PCR.
| Genes | Orientation | Primer sequences |
|---|---|---|
| GAPDH | Sense Antisense | 5’-GGA AGA TGG TGA TGG GAT TT-3’ 5’-TCT GAT TTG GTC GTA TTG GG-3’ |
| FOXP3 | Sense Antisense | 5’-GGC AGG CAA GAC AGT GGA A-3’ 5’-CAA GGA AAG GAG GAT GGA CG-3’ |
| T-bet | Sense Antisense | 5’-AAG ATA TGC GTG TTG GAA GC-3’ 5’-TGT GAC CCA GAT GAT TGT GC-3’ |
| GATA3 | Sense Antisense | 5’-TGA ACA AAT GAT TCG CCT A-3’ 5’-ATG AAG GAT GCC AAG AAG T-3’ |
| RORγt | Sense Antisense | 5’-GAG TGG GAG AAG TCA AAG ATG GA-3’ 5’-CTC AAA GCA GGA GCA ATG GAA-3’ |
IL-17A, IL-23, IL-4, IL-10, TGF-ß1, IFN-y and HBsAg concentrations in plasma were determined by conventional sandwich ELISA using detection and capture antibodies from BioLegend (IL-17A and IL-23; San Diego, CA, US) and Santa Cruz (IL-4, IL-10, TGF-ß1, and IFN-γ; CA, USA). ELISAs were carried out according to the respective manufacturers’ instructions. Standards and samples were run in duplicate.
Western blottingPatients liver biopsy tissues and normal liver tissues from liver transplant providers were dissolved in a RIPA buffer (150 mM NaCl, 1% sodium deoxycholate, 1% Triton X-100, 0.1% SDS, 10 mM Tris-HCl pH 7.2), separated by SDS-PAGE, and transferred onto nitrocellulose membranes (Millipore, USA). Primary antibodies against the following proteins were used: FOXP3 (Cell Signaling, USA), T-bet (Santa Cruz, USA), RORγt (Ambion, USA), GATA3 (Santa Cruz, USA), and GAPDH (Abcam, UK). Primary antibodies were detected by using appropriate horseradish peroxidase-conjugated secondary antibody. Blots were developed by the chemiluminescent detection system from Pierce (USA).
Statistical analysisStatistical analyses were performed with Statistical Product and Service Solutions (SPSS) Windows 14.0 Statistical Software (Chicago, IL, US). The data were expressed as mean ± standard error of the mean (SEM) or as median with range. Statistical significance was assessed with a twotailed Student’s t-test between two groups and by one-way ANOVA for comparisons among multiple groups.
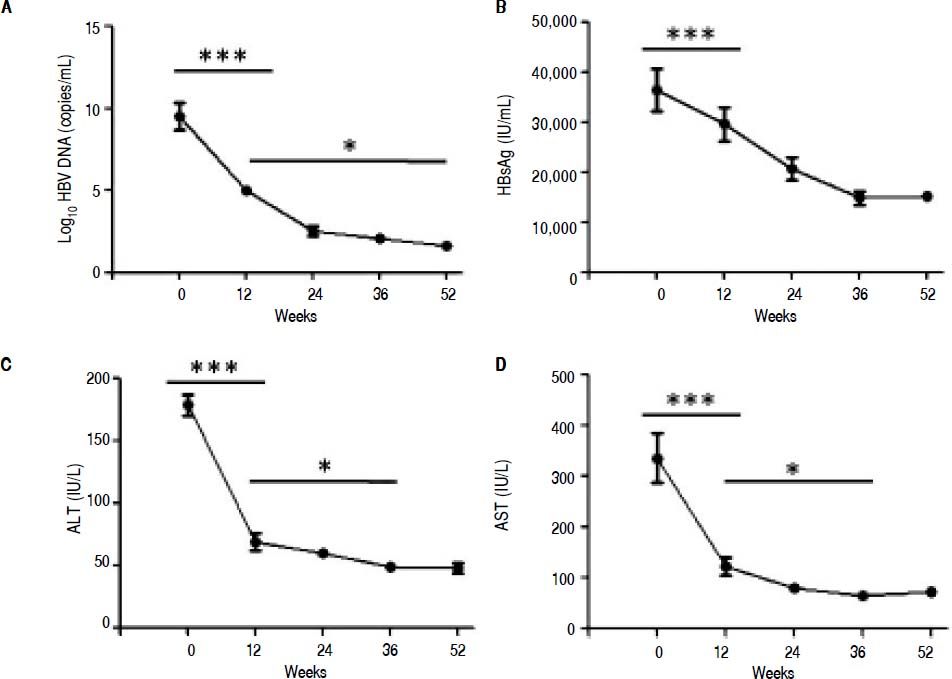
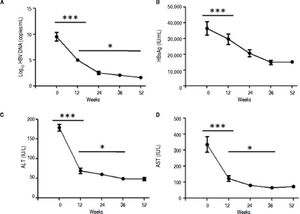
ResultsSerological and virological responses to entecavir therapy in CHB patientsThroughout the course of treatment, the viremia of all 30 patients showed declined to a low level (Figure 1). First of all, the HBV copy numbers sharply decreased after 12 weeks of treatment (Figure 1A). After that, we found steady decrease in the HBsAg concentration after 12 weeks of treatment till the end of experiment (Figure 1B). Finally, the plasma alanine aminotransferase (ALT) levels were found to be significantly lower compared to the levels at the start of 12 weeks treatment (Figure 1C). In addition, the plasma aspartate aminotransferase (AST) levels exhibited the same trend with ALT (Figure 1D).
Clinical characteristic of CHB patients after treatment with entecavir. Plasma samples were collected from 30 CHB patients treated with entecavir. HBV DNA level was detected by qPCR. Plasma HBsAg concentration was detected by ELISA. Plasma ALT and AST was measured by the CHEMIX-180 automated biochemistry analyzer (Japanese SYSMEX Corporation, Japan). Horizonta line indicates comparison of two groups. Asterisk indicates * P < 0.05 vs. baseline (0). ***P <0.001 vs. baseline (0).
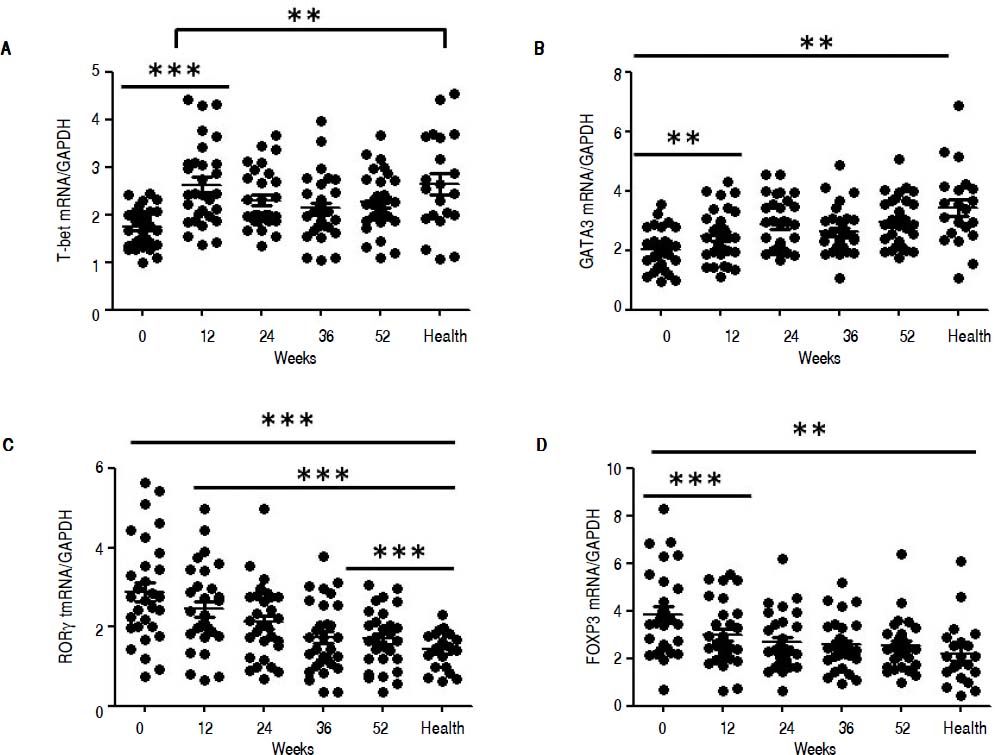
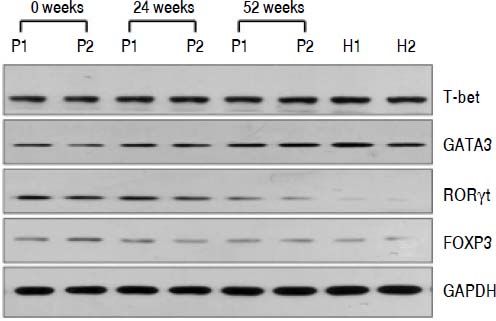
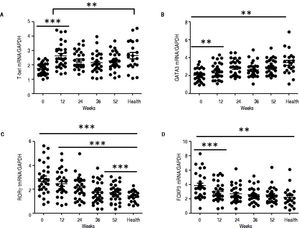
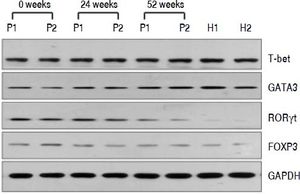
QRT-PCR results demonstrated that entecavir had different effects on the different CD4+ T-cell subtypes (Figure 2). All of the entecavir treatments did not produce significant changes in the expression of T-bet in CD4+ T cells of CHB patients (Figure 2A). However, the expression levels of GATA3 (the critical transcription factor of Th2 cells) in CD4+ T cells of CHB patients were remarkably lower than those in healthy volunteers throughout the course of treatment. But all of the entecavir treatments had markedly increased in the expression of GATA3 in CD4+ T cells of CHB patients (Figure 2B). The expression of RORγt (the critical transcription factor of Th17 cells) was found to decrease remarkably in CHB patients treated with entecavir, and the level reached close to the healthy volunteers throughout the course of treatment ( Figure 2C). The expression of FOXP3 (the critical transcription factor of Treg cells) decreased remarkably at 12 weeks of treatment. After 12 weeks, FOXP3 increased close to the expression of healthy volunteers (Figure 2D). In addition, entecavir effects on the different CD4+ T-cell subtypes were confirmed at the protein level. Western blotting results demonstrated that all their protein levels showed the same trend as the mRNA results as assessed by qRT-PCR (Figure 3). The protein level expression of T-bet was found to increase remarkably after 12 weeks of treatment, and it reached close to the protein level expression of healthy volunteers in the end of treatment (Figure 3).
Changes in the mRNA expression of the critical transcriptional factors for CD4+ T-cell subpopulations in CHB patients treated with entecavir. CD4+T cells from the healthy volunteers and CHB patients treated with entecavir were used to detect the relative mRNA expression levels of the CD4+ T-cell subtype-specific transcription factors by qPCR. **P < 0.01 and ***P < 0.001 vs. baseline (0).
Changes in the protein expression of the critical transcriptional factors for CD4+ T-cell subpopulations in CHB patients treated with entecavir. Liver biopsy tissues from CHB patients treated with entecavir were used to detect protein expression levels of the CD4+ T-cell subtype-specific transcription factors by western blotting. P: CHB patients. H: healthy person.
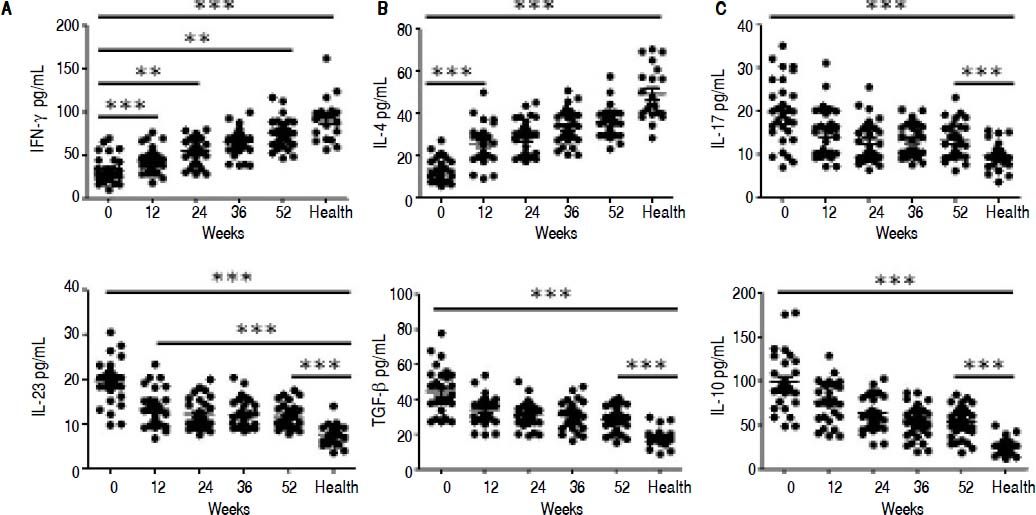
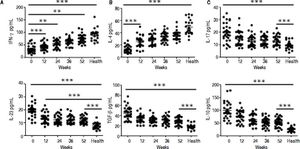
ELISA results demonstrated that entecavir has an impact on the expression of all the key cytokines in CD4+ T-cell subtypes (Figure 4). There was a gradual increase in the cytokines IFN-γ (Th1-related cytokine), and IL-4 (Th2-related cytokine) in CHB patients treated with entecavir (Figures 4A and 4B). In addition, entecavir treatment caused a gradual decrease in the cytokines IL-17 and IL-23 (Th17-related cytokines) and TGF-β1 and IL-10 (Treg-related cytokines) in CHB patients (Figures 4C-4F). All the detected cytokines were found to return to the normal levels, which was decreased but still higher at the end of experiment than it was at healthy volunteers (Figure 4).
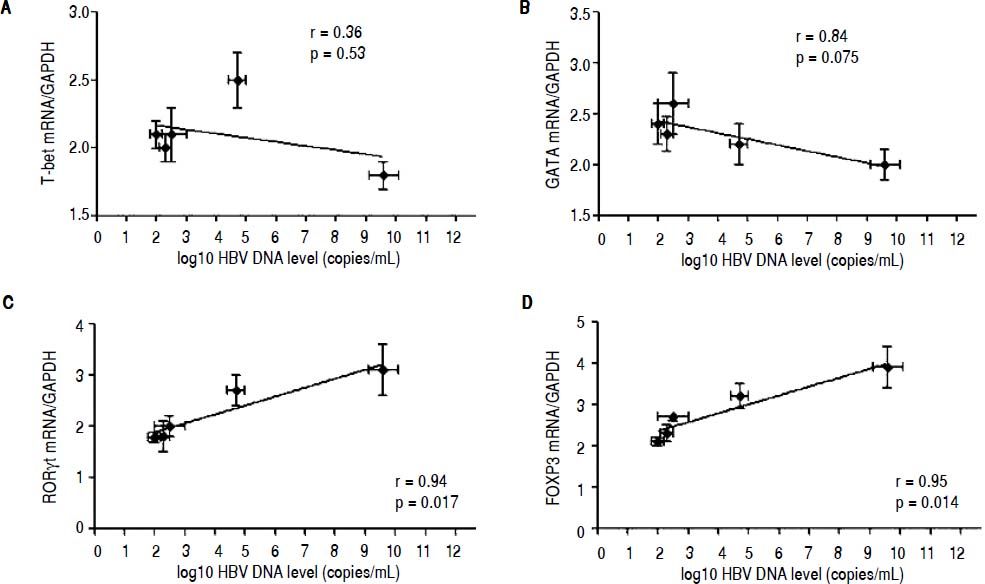
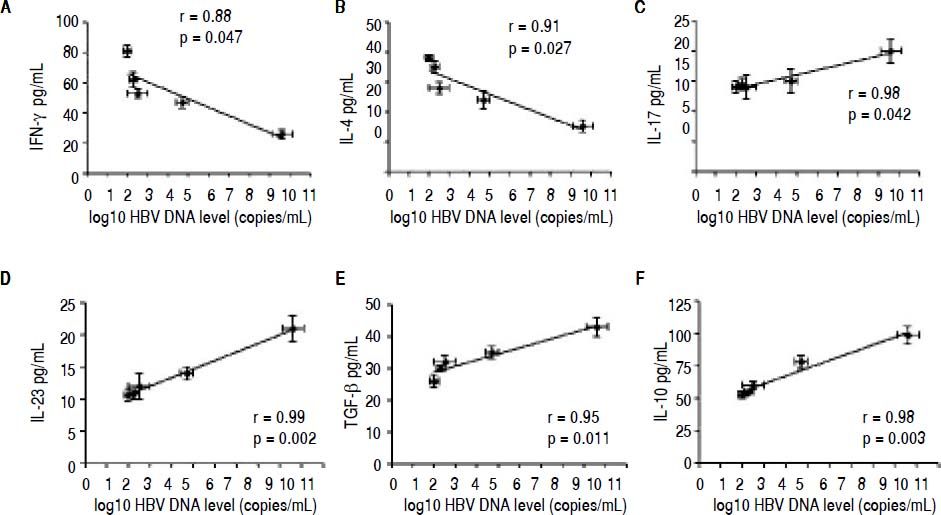
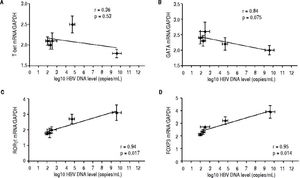
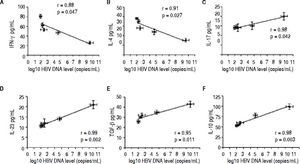
Correlation between the CD4 T-cell subtypes and HBV DNA levelsAbove all, the correlation between the critical transcription factor of each CD4+ T-cell subtype and HBV DNA levels was analyzed in the entecavir-treated CHB patients. Statistical analyses showed that a significant correlation was found between the expression of RORγt and HBV DNA levels in the entecavir-treated CHB patients. There was correlation between the expression of all transcription factors of each CD4+ T-cell subtype and HBV DNA levels in the entecavir-treated CHB patients, except for T-bet (Figure 5). Moreover, the correlation between signature cytokine levels of each of the CD4+ T-cell subtypes and HBV DNA levels was analyzed in the plasma of CHB patients. We found correlation between IFN-y, or IL-4, or TGF-β1 and HBV DNA levels. However, strong correlations were found between IL-17A, or IL-23, or IL-10 and HBV DNA levels (Figure 6).
The correlation between the critical transcription factor of each CD4+ T-cell subtype and HBV DNA levels. Expressions of the signature transcription factors for each of the CD4+ T-cell subtypes and HBV DNA level at various time points of treatment (baseline and weeks 12, 24, 36, and 52) are indicated on the plots by diamonds. Error bars indicate SEM.
The correlation between signature cytokine levels of each CD4+ T-cell subtype and HBV DNA levels. Expression of the signature cytokines for each of the CD4+ T-cell subtypes and HBV DNA level at various time points of treatment (baseline and weeks 12, 24, 36, and 52) are indicated on the plots by diamonds. Error bars indicate SEM.
In HBV infection, CD8+ T cells can recognize and clear virus-infected hepatocytes, reducing the levels of HBV.18–21 However, CD4+ T cells are required for the efficient development of effectors’ cytotoxic CD8+ T cells.6,22–24 In addition, CD4+ T cells can modulate the expression of the inflammatory cytokines in HBV infection,25 suggesting that CD4+ T cells play an important role in responding to HBV infection.
It is known that CD4+ T cells are a heterogeneous population, consisting of Th1, Th2, Th17, and Treg cells. However, which CD4+ T cell subsets play the most important role in CHB patients treated with entecavir is unclear. In order to investigate CD4+ T cell subsets, we measured cell type-specific transcription factors, which can directly reflect the cell frequency. As such, the expression levels of FOXP3, T-bet, RORyt, and GATA3 were detected in the mRNA and protein levels by qRT-PCR and western blotting, respectively. We found that there was a little effect on the Th1 cells of CHB patients, as the evidence showed that there was no significant change before and after treatment in liver biopsy tissue in protein level of T-bet. However, the Th1 signature cytokine, IFN-y, was found to a steady increase in the plasma of CHB patients treated with entecavir. This showed that the Th1 cell percentage was elevated by entecavir treatment, but those cells might move from the periphery to the liver tissue.26–27 This phenomenon needs more research to confirm.
Th2 cells may secrete cytokines IL-4, IL-5, IL-6 and IL-10, those involved in the humoral immune response in HBV infection. Both the expression of GATA3 (the Th2 critical transcription factor) and IL-4 (the Th2 signature cytokine) had a steady increase and reached the normal levels after the entecavir treatment. These results suggest that Th2 cells are dramatically increased by entecavir treatment in CHB patients. Such phenomena were also observed in the previous studies of CHB patients treated with telbivudine or adefovir dipivoxil.27,28 Because these drugs can inhibit HBV replication, they reduce the mobility of the virus particles, thereby enhancing the humoral immunity.
The previous studies have demonstrated that CHB patients have higher percentages of CD4+CD25+ regulatory T cells in their peripheral blood, compared to the normal control.7,29–31 Furthermore, those CD4+CD25+ regulatory T cells have immunosuppressive effects on HBV-specific T helper cells, which contribute to an impaired immune response against HBV, leading to chronic infection.7 Our data also showed that both the expression of FOXP3 (the Treg critical transcription factor) and TGF-β1 (the Treg signature cytokine) were found to be higher in CHB patients than in healthy controls. Moreover, entecavir treatment could rapidly reduce the FOXP3 expression in the CD4+T cells and liver biopsy tissues, and TGF-β1 and IL-10 in the plasma. Thus, entecavir treatment can reduce the frequency of Treg cells, leading to enhance the immune response to the HBV infection.
In accordance with Treg cells in CHB patients, Th17 cell frequency was also found to be higher in peripheral blood compared to the healthy control.32,33 Moreover, the elevated prevalence of Th17 cells is positively related with the increased plasma ALT levels in CHB patients.32 In this study, entecavir treatment can reduce the expression of RORyt in the PBMCs and liver biopsy tissues, and IL-17 and IL-23 in the plasma of CHB patients. Moreover, statistical analyses suggested that there was a strong correlation between the Th17 cell frequency and HBV DNA levels. These results indicated that Th17 cells were the subtypes most sensitive to entecavir-induced inhibition of viral replication, which was observed in a previous study of CHB patient with telbivudine.27
You, et al. had detected that T-cell subpopulations of chronic hepatitis B patients restored partially after entecavir treatment for 48 weeks.34 Zhang, et al. had confirmed the imbalance of Treg/Th17 cells was an important role for chronic hepatitis B patients during entecavir treatment.35 All those studies had confirmed the changes for T cells of chronic hepatitis B patients during long-term entecavir treatment. While we further examined each subgroup (Th1, Th2, Th17 and Treg cells) changes of CD4+Th cell, that helps us to understand the Naïve CD4+ cells differentiate into each subgroups during entecavir treatment.
ConclusionEntecavir treatment significantly decreased HBV DNA load, and therefore affected the CD4+ T-cell subsets in CHB patients. A dramatic decrease in the Th17 and Treg cell frequencies and expressions of their related cytokines were found in CHB patients with entecavir treatment. In contrast, entecavir treatment caused a significant increase in the Th2 cell frequencies and expressions of their related cytokines. Statistical analyses suggested that Th17 and Treg cells were the most sensitive subtypes to entecavir-induced inhibition of viral replication.
Author ContributionsZhan-Fei Tian and Zhong-Lan You contributed equally to this work.
Tian ZF, You ZL and Wang YM designed the research. Tian ZF, You ZL, Yi H and Kuang XM performed the operation. Wang YM, You ZL and Tian ZF collected the data. Tian ZF and You ZL wrote the paper. Wang YM reviewed the paper.
FundingThe work was supported in part by the State Key Project Specialized for Infectious Diseases (no. 2012ZX10002-004).
AcknowledgementThe authors thank all the subjects who participated in this study.