Background. The prevalence of two functional polymorphisms (rs1127354 and rs7270101) of the inosine triphosphatase (ITPA) gene associated with ribavirin-induced hemolytic anemia (RIHA) during antiviral therapy for hepatitis C virus (HCV) infection varies by ethnicity. In Mexico, the distribution of these polymorphisms among Native Amerindians (NA) and admixed population (Mestizos) is unknown. This study aimed to determine the prevalence of the ITPA polymorphisms among healthy NA and Mestizos, as well as in HCV patients from West Mexico.
Material and methods. In a cross-sectional study, 600 unrelated subjects (322 Mestizos, 100 NA, and 178 treatment-naïve, HCV-infected Mestizos patients) were enrolled. A medical history was registered. ITPA genotype was determined by Real-Time PCR. Fst-values and genetic relatedness between study and reference populations were assessed.
Results. The frequency of the risk genotypes rs1127354CC and rs7270101AA was higher among NA (98-100%) than in Mestizos (87-92.9%), (p < 0.05). The NA presented the highest prevalence of the rs1127354CC genotype reported worldwide. The Fst-values revealed a genetic relatedness among Mexican NA, South Americans and African populations (p > 0.05). The frequency of the predicted risk for RIHA was higher among NA (98%) than in Mestizos (80.5%) and HCV-infected patients (81.5%) (p < 0 .01). The CC/AA alleles were associated with lower values of total bilirubin, aspartate/alanine aminotransferases, and aspartate-to-plate-let-ratio-index score among HCV-patients.
Conclusion. A high prevalence of the ITPA polymorphisms associated with RIHA was found in Mexican NA. These polymorphisms could be a useful tool for evaluating potential adverse effects and the risk or benefit of antiviral therapy in Mexicans and other admixed populations.
Hepatitis C virus (HCV) infection is a significant health problem affecting 185 million individuals worldwide.1 While spontaneous clearance may occur in approximately 30% of individuals infected with HCV, the majority progress to chronic infection and are candidates for antiviral therapy.2 Also, once chronic infection is established it can advance to liver cirrhosis and hepatocellular carcino-ma.3
Before 2010, the standard of care for chronic HCV infection was based on pegylated-interferon alpha plus riba-virin (RBV); however, its effectivity was limited.4 As of 2011, with the approval of the new direct antiviral agents (DAAs) for chronic hepatitis C, such as boceprevir and te-laprevir among others, a higher efficacy has been achieved. These new DAAs target the nonstructural proteins involved in the HCV life cycle.5 In some cases, the DAAs offer an enhanced advantage when prescribed in conjunction with RBV. Thus, with the implementation of double or even triple therapy, the use of RBV will continue. RBV is a synthetic purine analogous effective against many DNA and RNA viruses, such as herpesviruses, poxvirus, influenza, measles, adenoviruses and hepatitis viruses.7 Unfortunately, RBV may not be easily tolerated because it has been associated with RBV-induced hemolytic anemia (RIHA). The development of RIHA during antiviral treatment against HCV infection is responsible for treatment discontinuation, dose reduction and consequently a decrease in the sustained virological response (SVR).8
The genetic locus responsible for the RBV-induced hemolytic anemia is found in the inosine triphosphatase (ITPA) gene, which encodes an enzyme with the same name.9 ITPA is a highly conserved enzyme that hydrolyzes deaminated purine (inosine) nucleoside triphosphates (ITP) into monophosphate and pyrophosphate deri-vates.9,10 As described by Felly, et al., the combination of two validated functional single nucleotide polymorphisms (SNPs) in the ITPA gene can predict its phosphatase (ITPase) activity.11 The ITPase activity predicted by the combination of two SNPs: rs1127354 (C > A) P32T, a missense variant located in exon 2 and the rs7270101 (A > C) a splicing-altering SNP located in the second intron is associated with RIHA.11 Specifically, the genotypes rs1127354AA and rs7270101CC lead to ITPase activity deficiency, a benign red cell enzymopathy characterized by an intense accumulation of ITP in erythrocytes and associated with protection against RIHA in HCV-infected pa-tients.12 By contrast, the genotypes rs1127354CC and rs7270101AA that lead to functional ITPase activity have been linked to RIHA, yet the molecular mechanism remains unknown. However, oxidative damage and erythro-cyte lysis have been related to the intracellular accumulation of pharmacologically active RBV.13
The prevalence of the ITPA polymorphisms and the predicted risk for RIHA has not been thoroughly studied across the American continent.14,15 As in most Latin American countries, the genetic structure of the Mexican population is an admixture of three paternal lineages consisting of Amerindian, European and African ancestry denoted as Mestizos, as well as conserved Native Amerindians (NA).16,17 Specifically, in Mexico, different degrees of admixture have been estimated among the Mestizos, while, on the other hand, there are nearly 15 million NA, who still maintain their inherited traditions.18 In consequence, a heterogeneous distribution of polymorphic genes associated with the clinical outcome and SVR of viral hepatitis, including the ITPA gene variants may be expected.19,20 Hence, the genotyping of these variants could be needed to evaluate the possibility of adverse effects and the risk or benefit of antiviral therapy, especially in patients that require extensive periods of treatment with ribavirin, even with the new DAAs. Therefore, the aim of this study was to determine the prevalence of the ITPA polymorphisms among healthy NA and Mestizos, as well as in treatment-naïve HCV patients from West Mexico.
Materials and MethodsStudy populationThis study was conducted at the Department of Molecular Biology in Medicine, Civil Hospital of Guadalajara “Fray Antonio Alcalde” in Guadalajara, Jalisco, Mexico. In this cross-sectional study, a total of 600 unrelated subjects from West Mexico were enrolled from January 2010 to December 2013. For the ITPA distribution study, 422 healthy individuals were included: 100 NA (50 Huicholes and 50 Nahuas) and 322 Mestizos (240 subjects from Guadalajara, Jalisco; 32 subjects from Villa Purificación, Jalisco and 50 subjects from Tepic, Nayarit). For the association study, 178 treatment-naïve HCV-infected Mestizos patients were included.
The NA were members of the Nahuas and Huicholes ethnic group, spoke their native language and had parents belonging to the ethnic group. The Mestizos subjects were defined as born in Mexico, which spoke Spanish, had Mexican parents and did not belong to any native group.16,21
The study protocol was approved by the Ethical Committee of the Hospital Civil of Guadalajara, Guadalajara, Jalisco Mexico and Hospital Israelita Albert Einstein, São Paulo, Brazil and was conducted in compliance with the ethical standards of the 2008 Declaration of Helsinki. All participants signed a written informed consent statement.
Clinical and biochemical evaluationThe healthy subjects were negative for anti-HCV antibodies. All HCV patients were treatment-naïve and negative for hepatitis B virus and human immunodeficiency virus at the time of enrollment. At this stage, a medical history questionnaire was used to register the amount of alcohol intake. Only patients consuming < 20 g of alcohol per occasion for women, and < 40 g alcohol per occasion for men (as recommended to prevent liver damage)22 were included in the study. Alcohol intake was calculated as previously reported.23 Anti-HCV antibodies were detected by a third-generation ELISA (AxSYM®, Abbott Laboratories, Illinois, USA). HCV viral load was measured by CO-BAS® AmpliPrep and COBAS® TaqMan® 48 HCV test (Roche Diagnostics, Pleasanton, CA, USA). Liver enzymes aspartate aminotransferase (AST), and alanine ami-notransferase (ALT) were determined by dry chemistry on a Vitros 250 analyzer (Ortho Clinical Diagnostics, Johnson & Johnson, Rochester, NY, USA). The aspartate aminotransferase-to-platelet ratio index (APRI) score was assessed to evaluate the degree of liver damage as previously reported.24
Isolation of genomic DNA and ITPA genotypingGenomic DNA was extracted from peripheral whole blood leukocytes by a modified salting-out method.25 The SNPs rs1127354 and rs7270101 were genotyped using a 5’ allelic discrimination method. A Real-Time PCR was carried out using predesigned TaqMan SNP Genotyping Assay (rs1127354 C_27465000_10 and rs7270101 C_29168507 _10, Applied Biosystems, Foster, CA, USA). An ABI 7500 Fast Real-Time thermocycler was used for PCR amplification following the standard conditions recommended by the manufacturer. The 7500 software (version 2.0.6.) automatically attributed the sample’s genotype. The correct genotype allocation was verified by positive and negative controls. Also, 20% of the samples were rerun, and 100% were concordant. The genotyping experiment was conducted in collaboration with the Albert Einstein Medicina Diagnóstica, Hospital Israelita Albert Einstein, São Paulo, SP, Brazil.
Prediction of risk for RIHAThe prediction of the risk or protection for RIHA was estimated according to the combined effect of the functional ITPA SNPs genotypes, as previously described.11,26 Briefly, high risk was rs1127354CC and rs7270101AA; moderate risk, rs1127354CC and rs7270101AC; mild risk, rs1127354CA and rs7270101AA or rs1127354CC and rs7270101CC; low risk, was rs1127354CA and rs7270101AC or rs1127354AA and rs7270101AA.
Genetic analysisThe genotype frequency of rs1127354 and rs7270101 SNPs was obtained by a direct counting method. The Har-dy-Weinberg equilibrium (HWE) expectation was assessed by the exact test. Genetic relatedness between populations based on ITPA SNPs was evaluated by pair-wise comparisons (exact test) and genetic distances (Fst) using Arlequin software version 3.0.27 These were additionally represented in a Multidimensional (MDS) scaling plot and in a Neighbor-Joining tree using SPSS software and FigTree version 1.4.2 for Windows (Institute of Evolutionary Biology, University of Edinburgh), respectively.
Statistical analysisThe categorical variables were expressed as frequency and were compared by chi-square or Fisher’s exact tests. Quantitative variables were expressed as median ± standard deviation (SD) and were compared with Student’s t-test. A p-value < 0.05 was considered as statistically significant. The statistical analysis was performed using SPSS version 21 for Windows (SPSS, Inc, Chicago, IL, USA).
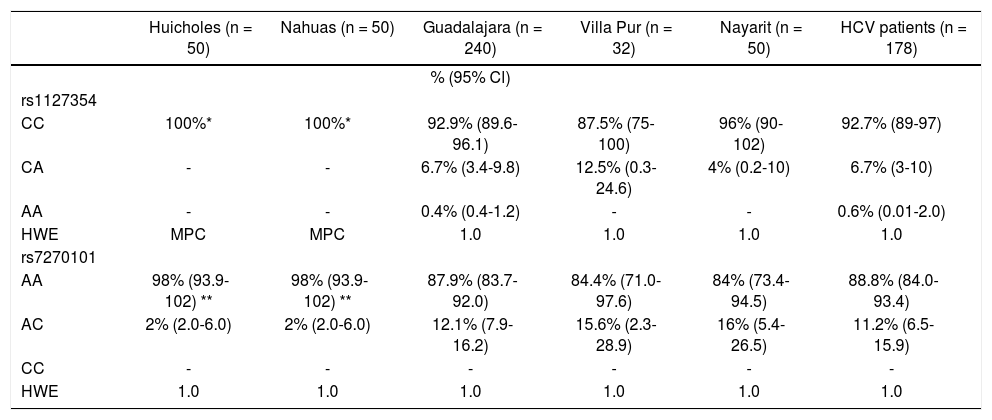
ResultsPrevalence of ITPA genotypes and comparative genetic analysisThe prevalence of the ITPA genotypes among the West Mexico’s populations is shown in Table 1. The genotype frequency of both SNPs differed between the NA and Mestizos. In regards to rs1127354, the wild-type CC genotype associated with risk of RIHA predominated in all the Mestizo populations (> 87.5%), and the Huicholes and Nahuas were monomorphic (CC = 100%). As for rs7270101, the AA genotype associated with risk of RIHA predominated among the Mestizo populations (> 84%). However, the Huicholes and Nahuas had the highest frequency (p < 0.05) compared to the Mestizos from Guadalajara, Villa Purificación, and Nayarit. All populations were in HWE for the two SNPs (p > 0.05).
Allelic and genotypic distribution of ITPA polymorphisms among Native Amerindians, Mestizos and HCV-infected patients
| Huicholes (n = 50) | Nahuas (n = 50) | Guadalajara (n = 240) | Villa Pur (n = 32) | Nayarit (n = 50) | HCV patients (n = 178) | |
|---|---|---|---|---|---|---|
| % (95% Cl) | ||||||
| rs1127354 | ||||||
| CC | 100%* | 100%* | 92.9% (89.6-96.1) | 87.5% (75-100) | 96% (90-102) | 92.7% (89-97) |
| CA | - | - | 6.7% (3.4-9.8) | 12.5% (0.3-24.6) | 4% (0.2-10) | 6.7% (3-10) |
| AA | - | - | 0.4% (0.4-1.2) | - | - | 0.6% (0.01-2.0) |
| HWE | MPC | MPC | 1.0 | 1.0 | 1.0 | 1.0 |
| rs7270101 | ||||||
| AA | 98% (93.9-102) ** | 98% (93.9-102) ** | 87.9% (83.7-92.0) | 84.4% (71.0-97.6) | 84% (73.4-94.5) | 88.8% (84.0-93.4) |
| AC | 2% (2.0-6.0) | 2% (2.0-6.0) | 12.1% (7.9-16.2) | 15.6% (2.3-28.9) | 16% (5.4-26.5) | 11.2% (6.5-15.9) |
| CC | - | - | - | - | - | - |
| HWE | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
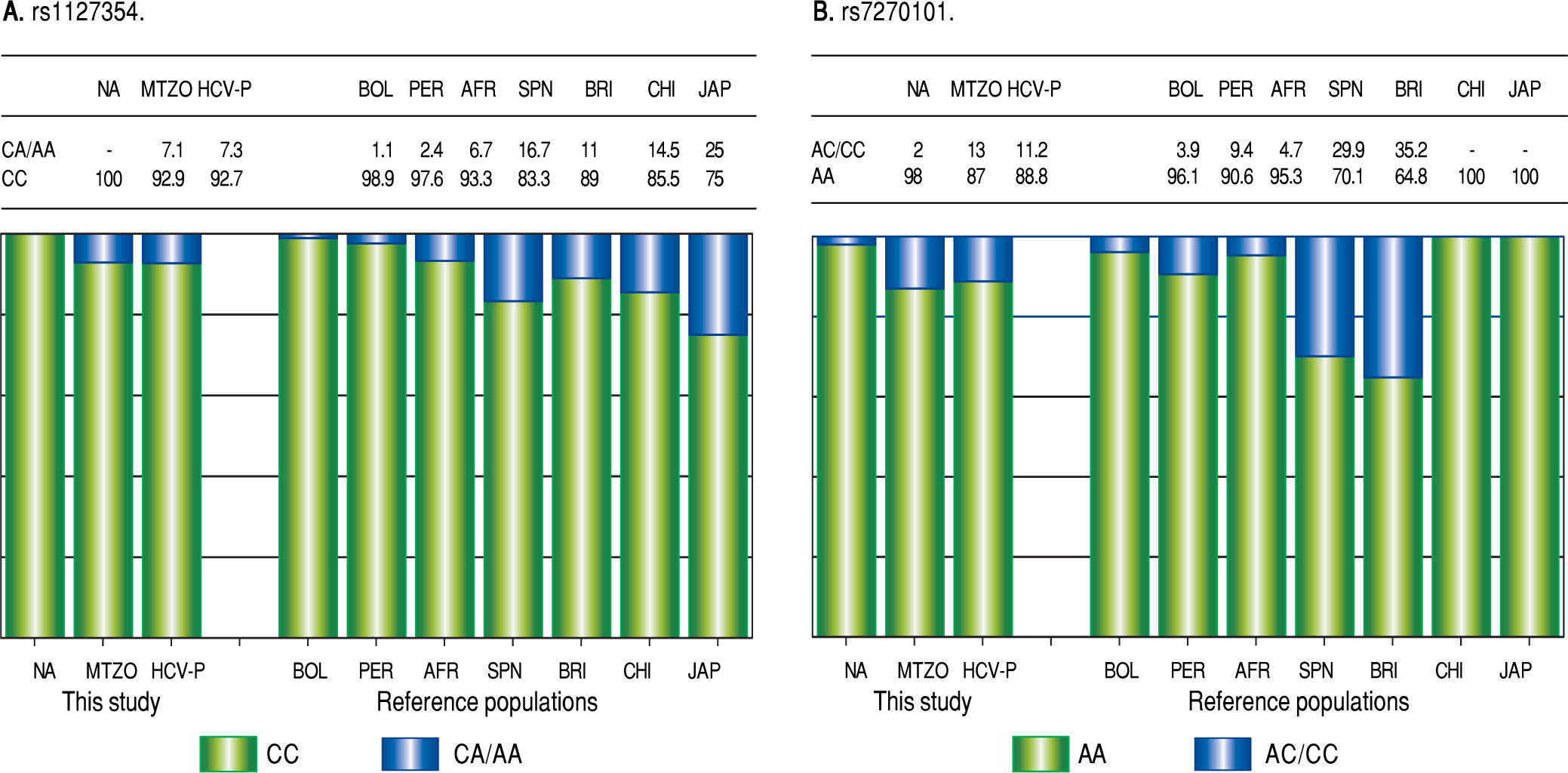
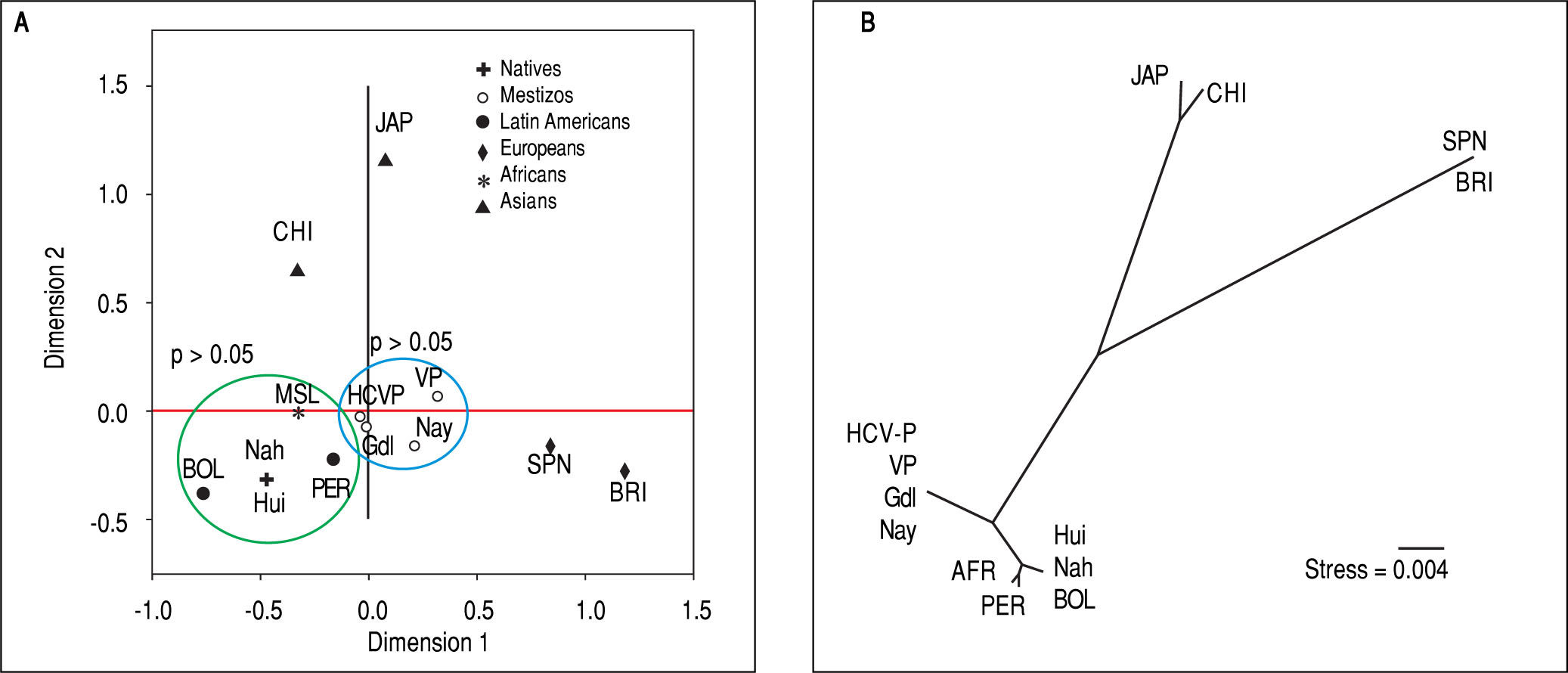
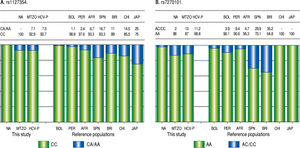
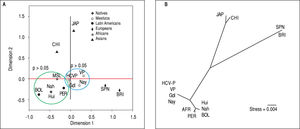
Due to the high prevalence of the risk ITPA genotypes among NA and Mestizo subjects, their prevalence was compared with reference populations with European, Asian, and African ancestry and other regions of Latin America reported in the 1000 Genomes Project28 (Figure 1). Furthermore, the genetic relatedness analysis between the study and reference populations (Bolivian, Peruvian, Caucasian, Asian and African) based on the two ITPA SNPs revealed two separate clusters. One cluster contained the NA from West Mexico, Bolivian and African individuals while the Mestizos formed a separate cluster and were poorly correlated to the Europeans and Asians (p > 0.05). (MDS plot and in Neighbor-Joining tree, Figure 2)
Prevalence of ITPA rs1127354 and rs7270101 polymorphisms in NA, Mestizos, HCV-infected patients compared with reference populations. A. The prevalence of the risk genotype (CC) was highest in NA than in all groups (p < 0.05) except in Bolivians (p = 0.296) and Peruvians (PER) (p = 0.407). While in Mestizos (MTZO) and HCV-infected patients (HCV-P) the prevalence was similar to all populations except Bolivia (BOL) (p = 0.002), China (CHI) (p = 0.021) and Japan (JAP) (p = 1.8 x 10-6). B. The prevalence of the risk genotype (AA) was highest in NA than in all groups (p < 0.05) except in BOL (p = 0.411), Africans (AFR) (p = 0.535), Japan and China (p = 0.464). While in MTZO and HCV-P, the prevalence was similar to PER and AFR but different to all others (p < 0.05).
Genetic relatedness based on the ITPA polymorphisms rs1127354 and rs7270101 between Mexican and reference populations represented on the multidimensional scaling (MDS) plot (A) and the Neighbor-Joining (NJ) tree (B). Population clustering depicted in the MDS plot is in agreement with spatial representation in the NJ tree. Hui, Huicholes. Nah, Nahuas. BOL, Bolivians. PER, Peruvians. Gdl, Guadalajara; Nay, Nayarit. VP, Villa Purificación. HCV-P, HCV-infected patients. CHI, China. JAP, Japan. SPN, Spain. BRI, British. MSL, Mende in Sierra Leone, Africa. AFR, Africans.
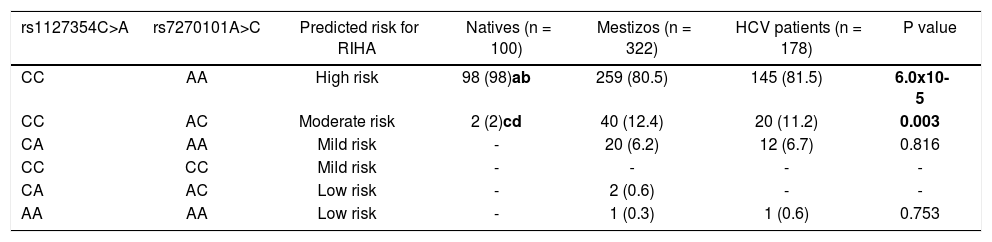
The prediction of the risk for RIHA was assessed using the combination of the two SNPs rs1127354 and rs7270101 (Table 2). Overall, NA subjects presented a high frequency of the risk genotypes (98%) associated with RIHA and were statistically significant compared to Mestizos subjects (80.5%) and HCV-infected patients (81.5%) (p=6 x 10-5). Meanwhile, up to 7.1% of the Mestizos and 7.3% of the HCV-infected patients had mild to low risk for RIHA.
Prediction of risk for RIHA in Native Amerindians, Mestizos and HCV patients
| rs1127354C>A | rs7270101A>C | Predicted risk for RIHA | Natives (n = 100) | Mestizos (n = 322) | HCV patients (n = 178) | P value |
|---|---|---|---|---|---|---|
| CC | AA | High risk | 98 (98)ab | 259 (80.5) | 145 (81.5) | 6.0x10-5 |
| CC | AC | Moderate risk | 2 (2)cd | 40 (12.4) | 20 (11.2) | 0.003 |
| CA | AA | Mild risk | - | 20 (6.2) | 12 (6.7) | 0.816 |
| CC | CC | Mild risk | - | - | - | - |
| CA | AC | Low risk | - | 2 (0.6) | - | - |
| AA | AA | Low risk | - | 1 (0.3) | 1 (0.6) | 0.753 |
Biochemical variables stratified by ITPA genotypes (CC/AA vs. all others) were evaluated in HCV-infected patients as shown in Table 3. The risk CC/AA genotypes were associated with lower levels of total bi-lirubin, AST, ALT and APRI score compared to patients with mild to low-risk genotypes (p < 0.05). No differences in hemoglobin and age were observed among these patients.
Biochemical profile of HCV-infected patients stratified by risk and non-risk ITPase genotypes
| Parameter | High risk CC/AA genotype (n = 145) | Non high-risk genotypes* (n = 33) | P value |
|---|---|---|---|
| Subjects (n, %) | 145 (81.5) | 33 (18.5) | - |
| Female (n, %) | 50 (39.1) | 8 (28.6) | 0.257 |
| Age (years) | 51.3 ± 11.9 | 50.1 ± 13.3 | 0.653 |
| Hemoglobin (g/dL) | 14.3 ± 1.7 | 14.03 ± 1.4 | 0.418 |
| AST (IU/L) | 54.6 ± 36.0 | 89.2 ± 103.2 | 0.004 |
| ALT (IU/L) | 62.5 ± 56.8 | 103.7 ± 120.1 | 0.011 |
| Platelets (x103/μL) | 188.2 ± 82.7 | 187.8 ± 88.8 | 0.959 |
| HCV load (x106 IU/mL) | 4.13 ± 9.38 | 4.35 ± 8.64 | 0.910 |
| APRI score | 0.85 ± 0.94 | 1.5 ± 1.7 | 0.014 |
| Total bilirubin (mg/dL) | 0.86 ± 0.51 | 1.3 ± 1.67 | 0.004 |
| Chronic Infection (n, %) | 84 (65.1) | 17 (60.7) | 0.659 |
In this study, we reported a high prevalence of ITPA polymorphisms and predicted high risk of RIHA among Mestizos, NA and HCV-infected patients residing in West Mexico. In this region, the Mestizos have mainly European ancestry followed by NA and in lesser extent African, as demonstrated by paternal lineages and maternal mtDNA haplogroups.16,29 However, in this study, the prevalence of the ITPA polymorphisms was higher in NA than in Mestizos and HCV-infected patients. Interestingly, the Mestizos and HCV-infected patients also carried the risk genotypes for RIHA, which may be due to the inheritance of the Amerindian component as shown by the comparative genetic analysis. These analyzes revealed that for the rs1127354 SNP, the Mestizos and HCV-infected patients were similar to Spanish and British subjects and different to Bolivian, Chinese and Japanese individuals (p < 0.05). In contrast, Huicholes and Nahuas had the highest frequency of the risk genotype (CC) compared to the European, African, and Asian population and they exhibited a similar frequency as in the Bolivian and Peruvian individuals (p = 0.296).15,28 These results are in concordance with previously described frequencies for ITPA in these populations from South America, exhibiting high NA ancestry.15 Also, these data are in agreement with the history of the peopling of Central and South America regions 15 000-25 000 years ago, which according to molecular analysis, share the same ancestors.30,31 Moreover, in regards to the rs7270101, the risk genotype (AA) had the highest frequency in NA from West Mexico, as well as Bolivians and surprisingly Asians. While, the Mestizos and HCV-infected patients were similar only to Peruvians and Africans. A possible explanation for this result is that NA have their origin in Asians founders who arrived via Beringia to the American continent.31 Also, rs1127354C and rs7270101A are considered both the ancestral alleles, which may be another reason they were almost fixed in NA and other ancestral populations.
In support of these hypotheses, both the MDS plot and the Neighbor-Joining tree showed the genetic relatedness between the NA from West Mexico, Bolivians, Peruvians, and Africans. However, the relatedness was not evident with the Asians subjects because the multidimensional representation was constructed using both SNPs and their frequencies were similar only in the rs7270101 polymorphism. Moreover, the Mestizos (including HCV-infected patients) were similar to themselves and occupied an intermediate position between NA groups and European populations (Spanish and British).
Given the reported consistency between the ITPA enzyme activity and the corresponding rs1127354 and rs7270101 ITPA alleles, these genetic variants have been proposed as predictors of drug toxicity and RIHA during HCV antiviral treatment in distinct populations,32,33 thus eliminating the need for measuring ITPA activity in affected patients.11,26. In this study, 81.5% of the Mestizo HCV patients may be at high risk of RIHA during antiviral therapy, and only 18.5% presented a predicted low risk. If we take into account that at least 700,000 Mexicans present active viremia, and many of them may have cirrosis,34 the use of DAAs plus RBV could be a common scheme of therapy in these patients; thus, RIHA and dose adjustment could compromise its effectiveness. However, it would be interesting to further investigate in admixed populations the ITPA genotype-phenotype correlation of these polymorphisms and others that have been reported.
Although, ITPA is highly expressed in liver, heart, sex glands, thyroid, and adrenal glands,35 no adverse effects associated with ITPase activity has been reported in these tis-sues.10 However, in this study, patients with the high-risk alleles showed significantly lower values of total bilirubin, ALT, AST, and APRI score compared to patients with moderate to low risk alleles. Interestingly, it has been documented that components of the bilirubin pathway may be altered during viral infection36 and may play an immune regulatory role in the outcome of HCV infection.37 Biliver-din is known to inhibit the HCV viral protease, whereas biliverdin reductase upregulation has been observed in chronically HCV-infected patients with an SVR to treatment relative to non-responding patients.37 Furthermore, the induction of heme oxygenese-1 in HCV infection has been shown to decrease viral replication, as well as protection against oxidative damage.38 On the other hand, the lack of elevated ALT and AST levels is consistent with retrospective studies of HCV-infected patients who revealed an association between the presence of symptoms, jaundice and spontaneous viral clearance status of serum polymerase chain reaction-negative individuals.39 Therefore, besides being at risk for RIHA, these patients may have alterations in the progression of HCV infection modulated by this particular clinical profile that requires further study.
Moreover, in conjunction with the ITPA polymorphisms, other genetic predictors for an antiviral response in chronic hepatitis C patients are the interleukin-28B (IL28B) and interferon lambda-4 (IFNL4) gene polymor-phisms.40,41 We have recently reported a genetic differentiation of the IL28B/IFNL4 haplotypes among Huicholes and Nahuas, who are carriers of the risk genotypes for HCV chronic infection and poor SVR.42 Therefore, if NA people were eventually infected by HCV, they could be at high risk of developing RIHA and/or may not respond to treatment. Likewise, the Mestizos, who presented a high prevalence of the risk alleles, could also have an adverse prognosis for treatment response. Thus, the characterization of ITPA and IL28B/IFNL4 that is currently being used as clinical markers of treatment response in Latin American countries such as Argentina and Brazil,43,44 could also be relevant in the Mexican population.45 Recently, the new DAAs have been incorporated into the health systems in Mexico. However, given the heterogeneous genetic background of the Mexican population for the aforementioned polymorphisms, it would be advisable to test patients to avoid poor responses to antiviral therapy.
Finally, the pharmacogenetic implications of the ITPA risk alleles are dependent on the drug therapy involved.46 These variants have been studied in the context of toxicity to 6-mercaptopurine and azathioprine in several diseases, including acute lymphocytic leukemia,47 inflammatory bowel disease,48 and transplant rejection.49 Thus, the results of this study could have an impact on the prediction of toxicity of drugs implicated in several diseases, as well as in hematological complications of viral infections treated with RBV.50
ConclusionThis study is the first to report the high prevalence of the ITPA risk alleles associated with RIHA in NA, Mestizos and treatment-naïve HCV-infected patients from West Mexico. These findings highlight the importance of pre-treatment characterization of the ITPA polymorphisms to evaluate potential adverse effects and the risk-benefit of antiviral therapy with RBV, especially in regions where NA ancestry prevails. Further investigations of these variants are necessary to assess the impact on the outcome of HCV antiviral therapy among admixed populations in Latin America and worldwide.
Abbreviations- •
HCV: hepatitis C virus.
- •
ITP: inosine triphosphate.
- •
ITPA: inosine triphosphatase.
- •
NA: native Amerindians.
- •
peg-IFN: pegylated interferon.
- •
RBV: ribavirin.
- •
RIHA: ribavirin-induced hemolytic anemia.
The study was supported by Promep-University of Guadalajara (UDG-CA-478) to AP and by the São Paulo Research Foundation (FAPESP)/CNPq (Grant 2010/ 10549-1) and the Hospital Israelita Albert Einstein, São Paulo, Brazil to JRRP.