Aiming to improve the anatomical resolution of electrical impedance tomography images, we developed a fuzzy model based on electrical impedance tomography’s high temporal resolution and on the functional pulmonary signals of perfusion and ventilation.
INTRODUCTIONElectrical impedance tomography images carry information about both ventilation and perfusion. However, these images are difficult to interpret because of insufficient anatomical resolution, such that it becomes almost impossible to distinguish the heart from the lungs.
METHODSElectrical impedance tomography data from an experimental animal model were collected during normal ventilation and apnea while an injection of hypertonic saline was administered. The fuzzy model was elaborated in three parts: a modeling of the heart, the pulmonary ventilation map and the pulmonary perfusion map. Image segmentation was performed using a threshold method, and a ventilation/perfusion map was generated.
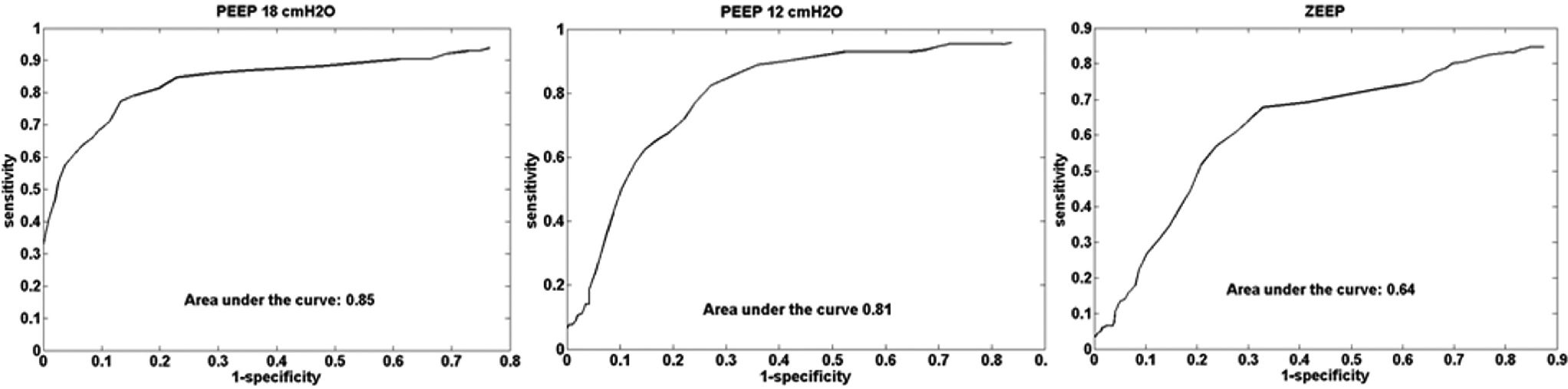
RESULTSElectrical impedance tomography images treated by the fuzzy model were compared with the hypertonic saline injection method and computed tomography scan images, presenting good results. The average accuracy index was 0.80 when comparing the fuzzy modeled lung maps and the computed tomography scan lung mask. The average ROC curve area comparing a saline injection image and a fuzzy modeled pulmonary perfusion image was 0.77.
DISCUSSIONThe innovative aspects of our work are the use of temporal information for the delineation of the heart structure and the use of two pulmonary functions for lung structure delineation. However, robustness of the method should be tested for the imaging of abnormal lung conditions.
CONCLUSIONSThese results showed the adequacy of the fuzzy approach in treating the anatomical resolution uncertainties in electrical impedance tomography images.
In an Intensive Care Unit (ICU), the functions of many organs are monitored using different devices: cardiac monitor, pulse oximeter and invasive arterial pressure cannula, among others. The lungs, despite the fact that they are vital organs, are usually not directly monitored at the bedside. The physiological functions of the lungs, necessary for maintaining gas exchange, are ventilation (air distribution) and perfusion (blood circulation). Electrical Impedance Tomography (EIT), a functional imaging method based on the distribution of electrical impedances within a volume (the human chest in this case), has the potential to show these pulmonary functions. EIT devices are small and portable and do not cause harm to the patient.1 Victorino et al. have been developing an EIT method to be used for bedside lung monitoring in the ICU. They have demonstrated that the variations in EIT impedance images of the lungs are very well correlated with the changes in the air content within a region of interest.2 However, the cyclical movement of blood through the pulmonary vessels also influences thoracic impedances. Consequently, at the least EIT images carry information about both ventilation and perfusion. Eyüboglu has demonstrated that it is possible to separate the thoracic impedance variations due to blood perfusion and those due to ventilation by using ECG-gated EIT images.3 Nevertheless, even after applying the ECG-gated method, the resultant images are difficult to interpret because of their insufficient anatomical resolution, and it is almost impossible to separate the heart from the lungs. In this paper we propose a fuzzy linguistic model for analyzing EIT images in order to identify and separate the heart from the lung regions. In addition, we propose a method to map, within the lung region, its two main functions, namely ventilation and perfusion.
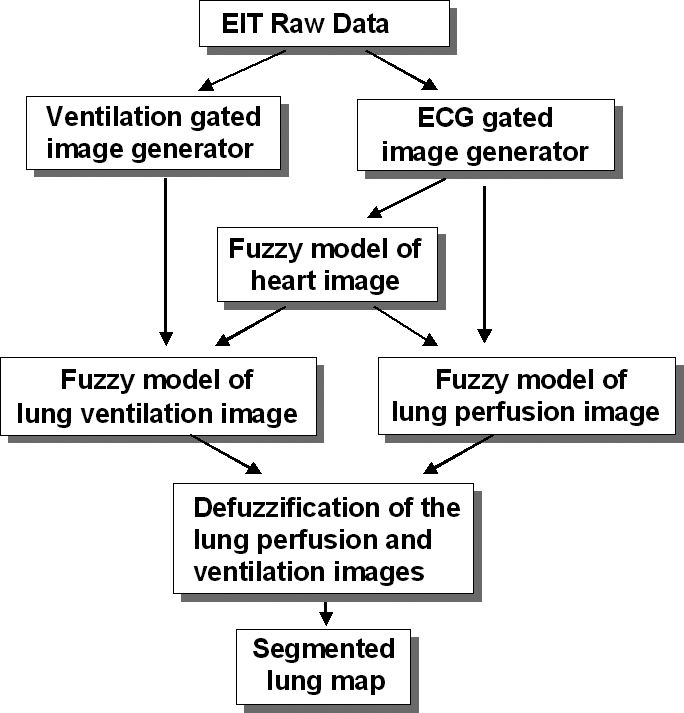
MATERIALS AND METHODSRecently, fuzzy set theory has been used to deal with uncertainties present in health sciences, and the results are very promising. Its application covers a wide range of subjects, from epidemiological studies to diagnosing system development.4–7 Our implementation of the EIT image treatment system employs the method of Mamdani and comprises software modules grouped in three steps: raw EIT data acquisition and image generation, fuzzy modeling and image segmentation (Figure 1).
A. EIT image generation module1) Raw EIT Data acquisitionThe experiments were performed on an anesthetized healthy pig lying in the supine position, because the purpose of this work was to establish a fuzzy model under a well-defined physiological condition. The animal was tracheostomized and maintained with a continuous infusion of anesthetics. Controlled mechanical ventilation was delivered with a Servo 300 A (Siemens, Sweden). For the injection of hypertonic saline, a catheter was placed into the superior vena cava next to the entrance to the right atrium. For the measurement of the pressure pattern of the cavities, a Swan Ganz catheter was also placed into the superior vena cava. ECG monitoring was performed using a Portal DX 2020 monitor (Dixtal, Brazil). Raw EIT data were acquired using an impedance tomography device based on the ENLIGHTER technology (Dixtal, Brazil), capable of producing 50 images per second. Thirty-two electrodes were placed circumferentially (equally spaced) around the thorax just below the level of the axilla. An electrical current of 5 mA was injected at 125 KHz through a pair of electrodes using an interleave one pattern. This means keeping one non-injecting electrode interposed between the injecting electrodes. Differential voltages were measured between the other non-injecting pair of electrodes with an interleave one pattern. Following this initial injection, the electrical current was then injected sequentially via the next pair of electrodes and repeated until all electrodes had served for current injection. The data for one complete cycle produced a so-called “frame,” and they were saved in a raw data file for later processing.
Ventilation was delivered by a mechanical ventilator in pressure control mode. The protocol had three steps of data collection characterized by the PEEP value: PEEP at 18 cm H2O, PEEP at 12 cm H2O and PEEP at 0 cm H2O (ZEEP). Each of these steps was composed of seven different phases of data collection: 1) the pulmonary embolism model, with occlusion and deflation of the Swan Ganz balloon; 2) the pulmonary artery occlusion pressure study; 3) the cardiac debit study; 4) the pulmonary artery pressure pattern study; 5) the left ventricle pressure pattern study; 6) the right atrium pressure pattern study; and 7) the superior vena cava pressure pattern study. In all of these 21 phases, apnea and saline infusion procedures were performed, resulting in 21 raw EIT data sets collected. In each phase of data collection, the Swan-Ganz distal pore was placed at the vessel region of interest, and the correspondent pressure curve was acquired, synchronously with EIT data. In off-line analysis, these pressure curves of each vessel and cardiac chamber were used only for qualitative comparison to the impedance time curve, with the purpose of characterizing the impedance time curve in each anatomic region. Respiratory rate was constant at 20 cycles per minute. Each raw EIT data set consisted of a first set of 10,000 frames during normal ventilation. Thereafter the ventilation of the animal was put on hold for a pre-defined period (apnea), while the pressure within the airways was maintained at the PEEP level. During this state of apnea, 5,000 images were acquired. Then, 5 ml of a hypertonic saline of 20% NaCl was injected quickly through the catheter, and another 5,000 more images were acquired.
2) Offline generation of ECG-gated image setFor each raw EIT data set, the synchronously recorded ECG waveforms were used to reconstruct a sequence of time-varying ECG-gated images. From the ECG waveform, a trigger pulse was produced at the rising edge of each R-wave. Following the detection of the R-wave, a block of frames was stored until the occurrence of the subsequent R-wave (one cardiac cycle). This process was repeated until 100 complete cardiac cycles were stored and one “mean cardiac cycle” could be generated from them. Further mean cardiac cycles were generated from subsequent data sets of one hundred cardiac cycles. This sequence of mean cycles was processed using an image construction algorithm, thus obtaining a series of ECG-gated images in which the effects of ventilation were attenuated. 21 ECG-gated image sets were generated.
3) Offline generation of ventilation-gated image setFor each raw EIT data set, the beginning of each respiratory cycle was used as the trigger signal in order to generate the ventilation-gated images. A block of frames between two subsequent trigger signals (1 respiratory cycle) was stored, and this process was repeated sequentially until 12 separate respiratory cycles were stored. Then one “mean respiratory cycle” was generated from each block of 12 cycles. This process was repeated for all subsequent data sets. The resultant sequence of mean respiratory cycles was processed using an image construction algorithm, thus obtaining a series of ventilation-gated images, in which the effects of lung perfusion were attenuated. Twenty-one ventilation-gated image sets were generated.
4) Off-line generation of saline injection image setFor each raw EIT data set, the saline injection images set were generated. In order to minimize the cardiac pulsating effect on the saline injection image series, a low pass filter was used, with the cut-off frequency set at 0.5 Hz. As a final result, 21 saline injection image sets were generated.
5) EIT image construction algorithmFor image construction, we used the black-box/sensitivity matrix algorithm developed by Lima and collaborators.8,9
6) Fuzzy model developmentAs explained above, the experiment was performed for three different PEEP values, covering the situation from zero to high PEEP. The fuzzy model was developed using the data of PEEP at 18 cm H2O, due to the assumption that the lungs are more uniform in terms of perfusion and ventilation distribution under these conditions. The model developed was applied to the data collected for PEEP at 12 and zero cm H2O.
B. Fuzzy model for EIT image treatmentEach EIT image is formed by a matrix containing 32x32 pixels. The fuzzy modeled image was obtained by running the model once for each pixel, requiring 1024 runs to form one modeled image. All fuzzy linguistic models developed for this study applied the Mamdani inference procedure and the center of area defuzzification method, and the models were based on expert experience in EIT chest image analysis.
1) Heart fuzzy modelThe fuzzy linguistic model for the heart has three antecedent variables in its propositions: normalized perfusion amplitude; normalized time delay (TD), obtained dividing each pixel’s TD value by the period of cardiac cycle; and pixel position. All of these were derived from ECG-gated images. The model also has one consequent variable: the possibility that the pixel carries the heart information (heart possibility). The pixel position is derived from the pixel order. The pixel of order 1 is located at the upper-left corner and the pixel of order 1024 is located at the lower-right corner of the image. Pixel orders from 1 to 512 belong to the anterior region of the image, while pixel orders from 513 to 1024 belong to the posterior region of the image.
2) Lung perfusion fuzzy modelThe lung perfusion linguistic fuzzy model has two antecedent variables - normalized perfusion amplitude and normalized heart possibility - as well as one consequent variable, the possibility that the pixel carries lung perfusion information. The defuzzified output obtained from running the heart possibility model was normalized in the interval [0,1] to obtain the normalized heart possibility. This fuzzy model is composed using nine inference rules.
3) Lung ventilation fuzzy modelAs in the perfusion lung model, the lung ventilation fuzzy model has two antecedent variables - normalized ventilation amplitude and normalized heart possibility - and one consequent variable, the possibility that the pixel carries lung ventilation information. The normalized ventilation amplitude was derived from ventilation-gated images and was selected from the ventilation-gated images of an image data segment comprising at least one respiratory cycle. Then, for each pixel, the amplitudes were calculated as the difference of maximum and minimum impedance values in the selected respiratory cycles. The pixel amplitudes were normalized in relation to the highest amplitude. This fuzzy model is also composed of nine inference rules.
C. Median fuzzy modeled heart imagesThe heart fuzzy model, as previously described and depicted in Figure 1, was run for each of the 21 ECG-gated image sets, totaling 21 modeled heart images. Afterwards, three median heart images were generated, one for each step of the protocol: PEEP at 18 cm H2O, 12 cm H2O and ZEEP. Each median heart image was calculated as the pixel-by-pixel median of the seven modeled heart images of each step of the protocol.
D. Median fuzzy modeled lung perfusion imagesThe lung perfusion model, as previously described, was run for each of the 21 ECG-gated image sets, totaling 21 modeled perfusion images. Afterwards, three median perfusion images were generated as the pixel-by-pixel median of the seven modeled perfusion images of each step of the protocol.
E. Median fuzzy modeled lung ventilation imagesThe lung ventilation fuzzy model, as previously described, was run for each of the 21 ventilation-gated image sets, totaling 21 modeled ventilation images. In a way analogous to perfusion images, three median ventilation images were generated, one for each step of the protocol,
F. Median saline injection lung imagesThe hypertonic saline solution acts as an EIT image contrast medium because it is much more conductive than blood, generating a much higher signal than physiological perfusion. During the data acquisition protocol, a set of images was made during apnea (without ventilation) in which a hypertonic saline solution (20% NaCl) was injected through a catheter inserted inside the superior vena cava. A series of images was made just after the injection. These images show the pattern of saline flow inside the chest: from the right atrium through the right ventricle, pulmonary arteries and lung vessels, until the recollection through the pulmonary veins to the left atrium. For each saline injection image set, experts in EIT images observationally selected a representative saline injection lung image, which was then used as the reference image for the lung perfusion images10 (Figure 2). Finally, 21 saline injection lung images were generated. Afterwards, three median saline lung images were generated as the pixel-by-pixel median of the seven saline lung images of each step of protocol.
G. Segmentation of modeled perfusion and ventilation imagesFor evaluation purposes, and in order to partition the modeled images into regions of practical interest, three segmented images were generated, one for each step of protocol. The method used for segmentation was the threshold of the median modeled images. The thresholds were heuristically determined: 0.31 for the median ventilation image and 0.28 for the median perfusion image. The heuristic used was the experimental search of threshold values in order to obtain a qualitative match of the outer lung contour of the total lung map and one CT-scan image of a pig, acquired in a different animal but under similar conditions of ventilation and axial level.
The segmentation process generated two maps for each step: one representing lung perfusion and the other representing lung ventilation. A segmented image was generated, one for each step, as the pixel by pixel overlay composition of the two previous maps. A total lung map was generated, for each step as the classical union of the two previous ones.
H. Fuzzy modeled image evaluationFor each step, the median perfusion image was compared with the median lung saline injection image, considered as the reference image (Figure 2). Two variables were calculated: a) sensitivity, defined as the number of pixels that belonged at the same time to the lung perfusion map and the reference image, divided by the number of pixels in the reference image; and b) specificity, defined as the number of pixels that, at the same time, did not belong to either the perfusion map or the reference image, divided by the total number of pixels that did not belong to the reference image. Both sensitivity and specificity, when closer to a value of one, indicate a better match between reference and target images. To generate the segmented reference image, a fixed threshold of 0.1 was used. Then, sensitivities and specificities were calculated for each perfusion map, with varying thresholds from 0.1 to 1, stepped by 0.05, in order to build the ROC curve.
I. Comparison of segmented lung and CT-scan lung imagesThe aim of image segmentation is to partition the image into several constituent components. In our study, the major components of interest are the lung perfusion map, lung ventilation map and heart region. Despite the difference in purposes of the two imaging methods, we chose the Computerized Tomography scan (CT-scan) image as the anatomic gold standard for comparison with the fuzzy modeled segmented image. The total lung map was compared with a CT-scan lung image of a pig, acquired in a different animal but under similar conditions of ventilation and axial level. The CT-scan lung image was sub-sampled to a 32x32 pixel lung map, and the accuracy indexes were calculated for each PEEP step. The accuracy index was defined as the proportion of true pixels in the 32x32 matrix; where true pixels were defined as both true positive pixels (defined as the number of pixels that belonged at the same time to the total lung map and to the CT-scan lung image) and true negative pixels (defined as the number of pixels that, at the same time, did not belong to either the total lung map or the CT-scan lung image). An accuracy index of 1 (100%) means that the modeled total lung map completely matched the CT-scan lung map.
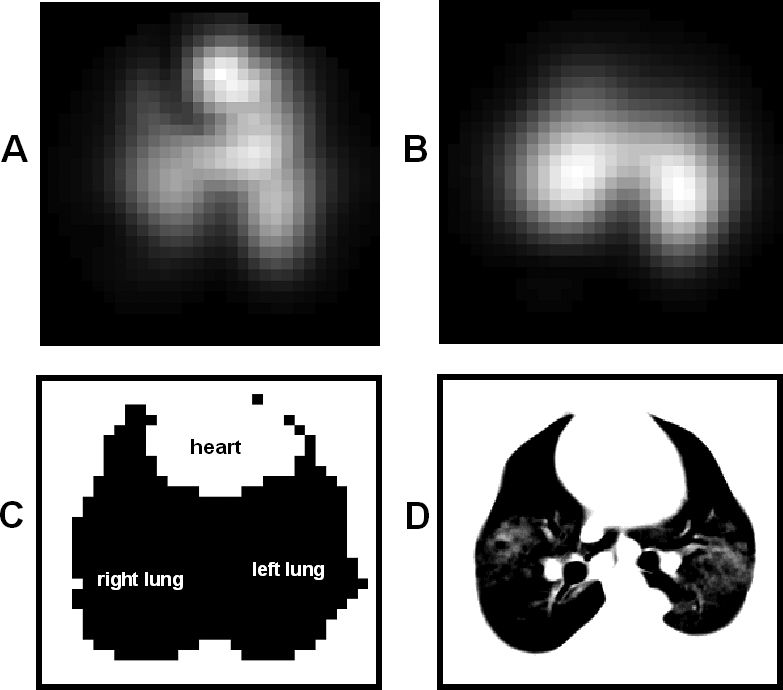
RESULTSIn all three PEEP steps, the fuzzy modeled heart images are identified in the anterior region (Figure 3, column a) as expected by the pig anatomy in the supine position. The lung perfusion possibility maps (Figure 3, column b) show a clear subtraction of the heart image from the original perfusion image. The same heart subtraction occurs with ventilation possibility images (Figure 3, column c).
Gray scale map of fuzzy modeling results: column a) median heart possibility map; column b) median lung perfusion possibility map; and column c) median lung ventilation possibility map. The lines represent the results for PEEP steps: from top to bottom, 18 cm H2O, 12 cm H2O and ZEEP
ROC curves (sensitivity vs. 1-specificity) were plotted comparing the lung perfusion image with the saline injection reference image (Figure 4), one for each PEEP step. The average area under the ROC curve was 0.77.
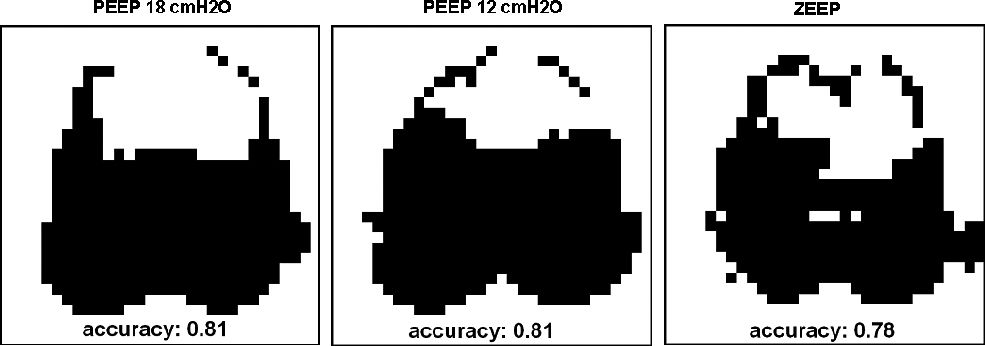
On the segmented lung map (Figure 5) the predominantly perfused regions (light gray) were found mainly for the lower regions of the lung. An increase in this region was noted, from the 18 cm H2O PEEP step to ZEEP. In ZEEP, the perfused region totally occupied the left lung region. The matched areas (dark gray) were concentrated in the middle region, and the predominantly ventilated regions appeared at the lung periphery. The heart region is clearly subtracted from the lung map. The comparison between the fuzzy modeled total lung maps (Figure 8) and the sub-sampled 32x32-pixel CT-scan lung map (Figure 7) resulted in an accuracy index of 0.81 for PEEP at 18 cm H2O, 0.81 for PEEP at 12 cm H2O and 0.78 for PEEP at zero cm H2O. In the CT-scan image there was a clear separation between the left and right lungs, but in the total lung map the lungs were presented as one single region, without left and right delineation. Figures 6-A and 6-B present the original EIT images of perfusion and ventilation, respectively, before the fuzzy analysis.
Total lung maps with respective accuracy indexes obtained from comparison to a sub-sampled 32x32 pixel lung map (figure 7). The maps from left to right correspond to PEEP steps at 18 cm H2O, 12 cm H2O and ZEEP, respectively
Lung map obtained by sub-sampling a CT-scan lung image (figure 6 - D) to a 32x32 pixel map
EIT is a medical imaging technology that has evolved from tracking pulmonary ventilation2 to the study of lung perfusion.3,10 However, combining the functional information contained in both ventilation and perfusion images to process clinically useful additional information remains a challenge. One important step towards achieving this objective is to construct a pulmonary mask that delineates the lung’s anatomic boundaries. Our fuzzy modeling of EIT images uses simultaneously acquired information from both lung ventilation and perfusion in order to determine the lung contour.
The heart fuzzy model proposed in this study clearly identified the heart region. When compared to a CT-scan image of the animal thorax, the modeled heart showed a good correspondence in both position and shape (Figures 3, column a, and 6-C). In this work we first identified the heart region and then, by heart subtraction, obtained the perfused and ventilated lung regions (Figures 3, column b and c). The anatomical a priori knowledge that the heart is normally situated in the anterior portion of the chest made it possible to use the pixel position as one of the antecedent variables of the fuzzy model. The use of pixel position resulted in a lower heart possibility in the posterior region of the image, as can be observed in Figure 3, column a. In contrast, the simple parameter of pixel position cannot be used to identify the lung regions, since pulmonary ventilation and perfusion occur in the anterior and posterior regions of the thorax at the same time.
The ROC curve (Figure 4) was plotted to evaluate the quality of the lung perfusion map in comparison to the reference method (Figure 2). The average area under the curve was 0.77, showing a good agreement between the two methods.
Comparing the segmented total EIT lung maps and the sub-sampled CT-scan lung map (Figure 7), the average accuracy index obtained was 0.80, with standard deviation of 0.02, showing a good similarity between the modeled and CT-scan lung maps. The low value of the standard deviation shows a high repeatability of the segmented total EIT lung map in different experimental conditions, in this case with varying PEEP values. On the CT-scan lung map, the right and left lung are separated by a central region not seen on the EIT-derived images. On the EIT segmented image the lungs are fused together in the middle of the thorax. We can point out one factor that may be responsible for this difference: the low spatial resolution of the EIT method. According to Thomas11, the observability of the central region is poor in the EIT method, compared to the peripheral regions.
The EIT image construction algorithm assumes that the measurement electrodes are placed equidistantly and on the same transverse plane of the thorax. In practice this may not always be true when individual electrodes are placed manually. Thus, there might be a considerable positioning error. Due to the lack of another applicable gold standard, we used EIT hypertonic saline injection images as an anatomical reference in this study. These reference images were produced by the same algorithm that generated the modeled fuzzy images and would therefore be subject to the same positioning errors.
The use of saline injection to map vascular perfusion is not new. Brown12 has already used it in humans to depict upper arm vessels, as well as the lung and heart, through the injection of saline from a dorsal hand vein. In his study, he was able to demonstrate not only venous perfusion but also an arterial perfusion at a later acquisition. Frerichs10 also proposed the use of saline injection as a contrast medium for the EIT method for diagnosis of pulmonary embolism. Therefore, saline injection may become a useful and reliable method of depicting heart and lung perfusion in critical patients, in part because it can be administered either by a central venous catheter or by peripheral access. In spite of the fact that the saline injection is innocuous in several clinical situations, it still requires an invasive method of venous access. The advantage of the fuzzy method would be its noninvasiveness, insofar as it uses only information gathered from time variations in physiological impedance.
Considering costs constraints as well as the fact that this was a preliminary study for proposing the use of fuzzy methods, we chose to use a single animal to derive our fuzzy model. In spite of that, our results show good reliability for the method. The fuzzy model is based entirely on EIT data, which are measurements of thoracic electrical impedance changes resulting from physiological phenomena. Electrical impedance is a physical property characteristic of each biological tissue. In similar tissue compositions, as in the thorax of different animals, one may expect a similar value for electrical impedance. The major determinant of thoracic electrical impedance variation is the amount of air inside the lungs, as already pointed out by Eyüboglu.3 In the experiments performed in this study; the PEEP was varied from zero to 18 cm H2O, which changes the amount of lung air content. Even with this change, which causes a major electrical impedance change, the fuzzy model showed good repeatability when compared with CT images, as shown by the low standard deviation of the calculated accuracy indexes.
Since the aim of this study was to develop an EIT imaging tool that is based on fuzzy models, the analyzed data were acquired only from a healthy pig. Thus, the robustness of the method should, in the future, also be tested in abnormal lung conditions.
The preliminary results of this study are encouraging, and future studies should be developed in order to answer questions regarding the clinical utility of the information presented by the fuzzy modeling.
CONCLUSIONSThe encouraging results of this study demonstrated the adequacy of the fuzzy modeling method for treatment of anatomical uncertainties in EIT images.
The model provided new lung structure delineation based on pulmonary functions not available in the original EIT images. The use of temporal information resulted in a significant enhancement of the spatial resolution of EIT images.