To determine the incidence of Mycobacterium tuberculosis complex and non-tuberculous mycobacterial isolates in the routine setting of a large general hospital using an “in-house” multiplex polymerase chain reaction method and to establish a paradigm for the definitive identification of mycobacteria isolated using semi-automated equipment.
METHODS:Established tests, including polymerase chain reaction restriction enzyme analysis, PNB, and NAP inhibition tests as the gold standard, showed 100% agreement with an IS6110/hsp65 multiplex polymerase chain reaction when used to identify stock strains (n = 117).
RESULTS:In a subsequent study, 8,790 clinical specimens producing 476 isolates were evaluated with multiplex PCR and also showed 100% agreement in identification using PRA-polymerase chain reaction as the gold standard. The application of this technique to routine analysis was demonstrated in this study. A method was established with the initial application of multiplex PCR for all positive liquid cultures and the subsequent identification of non-tuberculous mycobacteria by polymerase chain reaction restriction enzyme analysis. In total, 77% of isolates belonged to the Mycobacterium tuberculosis complex, and 23% were non-tuberculous mycobacteria.
CONCLUSIONS:Several non-tuberculous mycobacterial species were identified, primarily M. avium, but other potentially pathogenic species were also frequently observed, including M. fortuitum, M. abscessus, and M. kansasii. The expeditious communication of these data to the clinical staff was fundamental for the diagnosis of clinical cases. Even in settings where tuberculosis is of major importance, the incidence of non-tuberculous mycobacteria infection is substantial.
Mycobacterial infections are among the leading causes of disease in humans. In developing countries, the high incidence of tuberculosis overshadows the occurrence of non-tuberculous mycobacterial (NTM) infections, although this picture is changing. The incidence of NTM infections is increasing as a result of several conditions, such as the growing number of immunodeficient patients (1), the use of contaminated endoscopic medical devices, and cosmetic procedures (2). The correct identification of Mycobacterium is the mainstay for the clinical management of these patients.
The currently available commercial molecular biology tests designed for the direct identification of mycobacteria from clinical specimens have been employed (3-5), but they carry economic and methodological limitations.
Culture-based methods remain the gold standard for the specific diagnosis of these infections. Although Mycobacterium is readily isolated in solid or liquid media, the correct identification of the specimen is laborious and time consuming. There are few licensed assays that can accurately and quickly identify mycobacteria. The commercially available AccuProbe assay (Gene Probe, San Diego, California) and the polymerase chain reaction (PCR)-based reverse hybridization Inno-LiPA assay (Innogenetics, Ghent, Belgium) (6) are expensive and difficult to employ in resource-limited institutions.
Recently, PCR and PCR-linked in-house methods have been used for the rapid detection and differentiation of MTC and NTM in routine diagnostic laboratories. Multiplex PCR, which targets many different genes simultaneously, has been used for this goal (7-12). The incidence of MTC and NTM was determined in this work using an “in-house” method targeting a specific sequence in MTC organisms (IS6110) and the hsp65 gene found in both MTC and NTM. A method could thus be established with the initial definitive identification of the isolate using multiplex PCR followed by PRA-PCR as the standard species-level identification test.
MATERIALS AND METHODSThe study was performed at the Molecular Biology and Bacterial Pathogenesis Laboratory of the Clinical Medical Department in the Faculty of Medical Sciences in the State University of Campinas (Brazil). This study comprised two distinct studies. The first was designed to compare a simple PCR-based identification test (hsp65/IS6110 multiplex PCR) with reference tests (PNB and NAP) using a set of previously identified MTC and NTM strains. The second was intended to identify mycobacteria grown with the aid of a semi-automated culture system (MGIT 960) in the routine setting of a clinical laboratory using the previously tested hsp65/IS6110 multiplex PCR. PRA-PCR was used as the gold standard for both parts of the study.
Mycobacterial StrainsStudy one – Stock strains that were previously identified by PRA-PCR and recovered from clinical specimens obtained from patients who were previously admitted to the hospital were as follows: M. tuberculosis (n = 91), M. abscessus (n = 2), M. avium (n = 6), M. gordonae (n = 13), and other NTM (n = 2). The strains were preserved in Lowenstein-Jensen (LJ) media and transferred to MGIT bottles or fresh Lowenstein-Jensen media for further tests.
Study two – The study was designed to last from January 2009 until July 2010. All clinical material sent to the laboratory was processed and inoculated into MGIT bottles. All mycobacteria grown were identified with multiplex PCR. Samples of MTC and all NTM isolates that were previously differentiated by hsp65/IS6110 multiplex PCR were identified using PRA-PCR.
Clinical Specimens – The specimens were processed according to standard procedures already in use at the laboratory (13). All materials were handled in a class II type B2 laminar flow biological safety cabinet.
Culture – Potentially contaminated specimens were processed by the modified Petroff method (13), as recommended by the MGIT manufacturer. Other non-contaminated specimens, such as tissue fragments or sterile biologic fluids, were directly inoculated into culture tubes. All specimens were processed within 24 hours. Smears were stained by the Ziehl-Neelsen method. A total of 0.5 mL of processed specimen was added to a BACTEC MGIT 960 culture tube. For the first part of the study, PNB-supplemented Lowenstein-Jensen media slants were inoculated with the aid of a loop.
IdentificationMultiplex PCR - Mycobacterial DNA was extracted from MGIT culture media by centrifuging 0.5 mL of the liquid culture media. The resulting pellet was resuspended in sterile distilled water. The suspension was then heated once at 80°C for 20 min. Multiplex primers were designed to amplify sequences of both the hsp65 gene and the IS6110 repetitive element. The primers used in the study were the following: TB11 (hsp65) 5’-ACCAACGATGGTGTGTCCAT-3’; TB12 (hsp65) 5’-CTTGTCGAACCGCATACCCT-3’; TB284 (IS6110) 5’-GGACAACGCCGAATTGCG-3’; and TB850 (IS6110) 5’-TAGGCGTCGGTGACAAAGGCCAC-3’. Amplification was performed in a DNA thermal cycler (GeneAmp PCR 9600; Perkin-Elmer, USA). A 5 μL aliquot of each DNA sample was used in a total volume of 50 μL PCR mixture containing 10 mM Tris-HCl (pH 8.8), 50 mM KCl, 1.5 mM MgCl2, 0.1% Triton X-100, 0.2 mM dNTPs, optimal amounts of primers (12.5 - 25.0 pmol), and 1 U of Taq DNA polymerase. The multiplex PCR conditions were optimized as follows: one cycle of denaturation at 95°C for 3 min; 30 cycles consisting of denaturation at 95°C for 20 sec, annealing at 65°C for 30 sec, and extension at 72°C for 30 sec; and a final cycle of extension at 72°C for 7 min. Electrophoretic separation of each PCR product (10 μL) was performed in 2% agarose gel slabs in Tris-borate buffer [0.5×TBE; 0.04 M Tris-borate (pH 8.4), 1 mM EDTA] at 100 V for 30 min. The gels were stained with SYBR Green (Molecular Probes, Oregon, USA). Reference strains were run as positive controls (M. tuberculosis H37Rv; M. avium), and deionized water was used as the negative control. For comparison of product lengths, 1 μg of 100 bp ladder (Bio-Rad, USA) was used as a molecular size marker.
PRA-PCR - PCR restriction enzyme analysis of the hsp65 gene (PRA-hsp65) was performed according to previous reports (14,15). Briefly, an aliquot (5 μL) of each template DNA was used in 50 μL of PCR mixture containing 10 mM Tris-HCl (pH 8.8), 50 mM KCl, 1.5 mM MgCl2, 0.1% Triton X-100, 0.2 mM dNTPs, 12.5 pmol of each primer (TB11 and TB12), and 2 U of DNA polymerase. The amplification was performed as follows: one cycle of denaturation at 95°C for 5 min; 45 cycles consisting of denaturation at 94°C for 1 min, annealing at 62°C for 1 min, and extension at 72°C for 1 min; and a final cycle of extension at 72°C for 7 min. For BstEII (Promega, USA) and HaeIII (Promega, USA) enzyme digestions, 10 μL of each PCR product was added to a mixture containing 1 μL (20 U) of each enzyme, 3 μL of restriction buffer, and 16 μL of deionized water. The mixture was incubated for 60 min at 60°C for BstEII enzyme digestion and 37°C for HaeIII enzyme digestion. The electrophoretic separation of digested products was performed for 60 min at 100 V on 3% agarose gel slabs, which were imaged under UV light after staining with SYBR Green (Molecular Probes, Oregon, USA). Isolates were identified using the PRA algorithm as described previously.
PNB was incorporated into LJ medium or BACTEC MGIT960 7H9 medium to a final concentration of 500 μg/mL (16). Control growth tubes contained plain LJ medium and BACTEC MGIT960 7H9 medium without inhibitory substances. Inocula were prepared from cultures grown on Löwenstein-Jensen medium. A few colonies were emulsified in flasks containing glass beads and 2 mL of sterile distilled water, allowing the turbidity to be greater than the McFarland scale N° 1 standard. The resulting suspension was left to stand for 15 min to allow larger clumps to settle; 1 mL of the supernatant was then transferred to another tube, in which the turbidity level was adjusted to McFarland scale N° 1. This bacterial suspension was used as the working suspension.
The NAP test was performed in a BACTEC460 radiometric device. The isolate was incubated into 12B bottles to a growth index (GI) of 50 to 100. An aliquot of 1 mL was then transferred to a new 12B bottle (BACTEC460-radiometric) containing an impregnated disk with 5 μg of NAP. The GI of both bottles was recorded and compared every day for the next 6-7 days.
RESULTSAssessment of hsp65/IS6110 multiplex PCR with previously identified Mycobacterium strains - This part of the study was designed to compare traditional identification tests with hsp65/IS6110 multiplex PCR. The hsp65 and IS6110 sequences were easily and specifically amplified from DNA extracted with heat lysis from MGIT liquid culture. Identification with multiplex PCR was simple and more convenient than other tests conducted in this study, which demanded more time and more expensive materials. Initially, no amplification was obtained when we tried to perform PCR with crude lysates from MGIT aliquots. However, amplification was successful when the lysates were centrifuged and the resulting pellet resuspended in distilled sterile water.
There was 100% agreement between identification with PNB using either LJ media or MGIT and hsp65/IS6110 multiplex PCR. There was no difference between strains grown in LJ media and MGIT. The NAP test misidentified 6 cases. The results were consistent among all NTM strains and MTC strains tested (Table 1).
Distribution of mycobacterial isolates according to the identification test performed in the validation study.
| Multiplex PCR | NAP/BACTEC460 | PNB/LJ | PNB/MGIT | |||||
|---|---|---|---|---|---|---|---|---|
| PRA - PCR | MTC | NTM | MTC | NTM | MTC | NTM | MTC | NTM |
| MTC | 91 | 0 | 85 | 0 | 91 | 0 | 91 | 0 |
| NTM | 0 | 26 | 6 | 26 | 0 | 26 | 0 | 26 |
| Total | 91 | 26 | 91 | 26 | 91 | 26 | 91 | 26 |
NAP/BACTEC460 – NAP added to a 12B bottle; PNB/LJ – PNB added to LJ media; PNB/MGIT - PNB added to an MGIT bottle. MTC – Mycobacterium tuberculosis complex. NTM – Non-tuberculous mycobacteria.
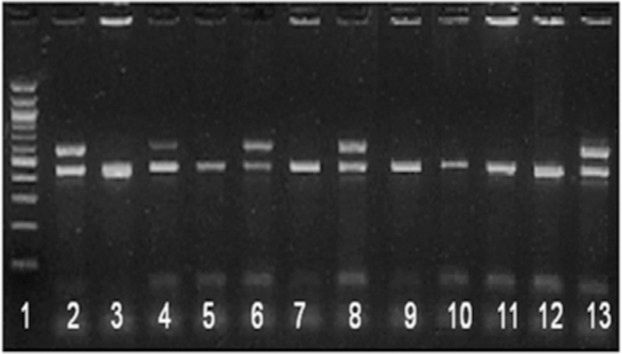
Application of hsp65/IS6110 multiplex PCR in routine settings - During the study period, 8,790 clinical specimens were consecutively received for culture, and 476 isolates were grown. All positive bottles were enrolled in the study and submitted to identification with hsp65/IS6110 multiplex PCR. In total, 367 (77%) isolates were identified as M. tuberculosis complex, and 109 (23%) were identified as NTM. For 19 isolates, amplification was not possible, or multiple bands were obtained (Table 2). Figure 1 shows the amplification products of some isolates. A total (n = 130) of 46 MTC isolates and 84 NTM, as identified by multiplex PCR, were identified using PRA-PCR. Respiratory secretions, sputum and broncho-alveolar lavage were the main sources of the isolates. M. tuberculosis was recovered from the sputum, lymph nodes, pleural fluid, urine, and skin. NTM were isolated from the sputum, blood, bone marrow, pleural fluid, gastric lavage, broncho-alveolar lavage, feces, and lymph nodes (Table 3).
Number of culture requests, MTC isolates and NTM isolates, as identified by multiplex PCR, during the study period.
| Year | Culture Requests | MTC | NTM | ND |
|---|---|---|---|---|
| 2009 | 5262 | 192 | 66 | 11 |
| 2010 | 3528 | 175 | 43 | 8 |
| Total | 8790 | 367 | 109 | 19 |
MTC – Mycobacterium tuberculosis complex. NTM – Non-tuberculous mycobacteria. ND – Not determined.
Distribution of mycobacterial species identified by PRA-PCR in the routine setting.
| Species | Clinical specimen | N |
|---|---|---|
| M. alvei | Broncho-alveolar lavage (BAL) | 1 |
| M. kubicae | Gastric lavage | 1 |
| M. lentiflavum | Sputum | 1 |
| M. mucogenicum | Sputum | 1 |
| M. sherrisii | Sputum | 1 |
| M. simiae/M. lentiflavum | Sputum | 1 |
| M. chelonae | Sputum | 2 |
| M. intracellulare | Sputum | 2 |
| M. kansasii | Lymph nodes, Sputum | 2 |
| M. parmense | Sputum, Pleural fluid | 2 |
| M. szulgai | Sputum, Lymph nodes | 2 |
| M. xenopi | Sputum | 2 |
| M. flavescens | Sputum | 3 |
| M. intracellulare/M. chimerae | Sputum | 3 |
| M. gordonae | Gastric lavage, Sputum | 5 |
| M. kansasii/M. branderi | Lymph nodes | 6 |
| M. abscessus | Sputum, Blood, BAL | 8 |
| M. fortuitum | Gastric lavage, Sputum | 8 |
| M. avium | Sputum, BAL, Bone marrow, Feces, Blood | 33 |
| M. tuberculosis complex | 46 |
There was 100% agreement between multiplex PCR and PRA-PCR in the samples studied. All strains that were previously identified as M. tuberculosis or NTM were confirmed by the reference test. Among NTM, 19 different species were identified, primarily M. avium as expected, but other potentially pathogenic species were frequently observed, including M. fortuitum, M. abscessus, and M. kansasii (Table 3). The unusual mycobacteria found in this study, including M. alvei, M. kubicae, M. sherrisii, and M. parmense, identified by PRA-PCR, require further confirmation by sequencing.
DISCUSSIONMycobacteria isolated in clinical settings require fast identification. The effective treatment of diseases caused by such agents differs among species and has to be instituted promptly. The clinical laboratory is essential in providing this information. Most isolates involved in human disease and reported in the literature belong to the M. tuberculosis complex or MAC, but recently, many other species have been described in association with hospital infections or cosmetic procedures (2).
In settings where tuberculosis is a major problem, specific diagnostic methods for the rapid detection and differentiation of NTM infections from TB are of major clinical importance. Identification based on the use of chemical inhibitors of growth, such as PNB, is reliable but still has disadvantages because it is time consuming, expensive, and can eventually yield false results, inhibiting M. kansasii and M. marinum growth (16,17).
When using methods based on mycobacterial genomic amplification, the choice of the target is fundamental for species or genus identification studies. Insertion sequence 6110 is exclusively found in members of the M. tuberculosis complex and has been employed in studies worldwide (18,19). Eventually, the amplification of this target may yield false-negative results when performed in some geographical areas (20) because the copy number of this sequence in M. tuberculosis isolates varies by up to 25 worldwide. Molecular epidemiology and other identification studies in our region show the universal presence of this sequence in M. tuberculosis strains, making it a valuable target for identification (21,11).
In this work, two sets of primers targeting IS6110, which is exclusively found in Mycobacterium tuberculosis complex members, and the genus-specific hsp65 gene were conjugated in a multiplex PCR to differentiate MTC from NTM. This method proved to be very sensitive and specific when tested against reference identification methods. The usefulness of this multiplex PCR was further evident in a clinical setting in the analysis of a large number of clinical specimens; many diverse mycobacterial species, as identified by PRA-PCR, could be separated from members of the M. tuberculosis complex. No false-negative results were obtained. All members of the M. tuberculosis complex identified by multiplex PCR were confirmed by the reference method. The same result was also found for non-tuberculous mycobacteria (NTM). When used in the routine laboratory setting, the test was very sensitive and specific, confirming the results of the preliminary study. A significant number of non-tuberculous mycobacteria were identified from clinical materials, accounting for 23% of all isolates. The large number of different species of NTM observed and the fast communication of these data to the clinical staff were fundamental for the diagnosis of clinical cases.
NTM isolates have been reported with an increasing frequency worldwide. This could be due to widely available sensitive culture methods and identification techniques, aging, immunosuppression or the inadequate sterilization of surgical devices.
A particular geographical distribution of NTM species has been reported. Rapidly growing mycobacteria (RGM) are the most frequently isolated mycobacterial group in Shanghai, as reported by Hong-xiu et al. (22), followed by MAC strains. MAC is not as frequently isolated in AIDS patients in Africa compared with other occidental countries (23,24), where M. fortuitum, M. kansasii, and M. xenopi are primarily found. The identification of NTM in clinical specimens from Brazil, mostly from regional reference laboratories, has been reported (25,26), and M. kansasii, M. avium complex, M. gordonii, and M. fortuitum are the most frequently observed. However, these reports are based on clinical specimens sent to the reference laboratory based on a distinct strict criterion, not on universally cultivated material, as we have studied. MAC strains, M. fortuitum, M. kansasii, and M. abscessus were most frequently isolated in our work.
The high sensitivity and specificity of the genomic targets used in this study have also been demonstrated in similar studies by others. When used to detect and identify reference strains of M. tuberculosis, M. fortuitum, M. smegmatis, M. avium, and M. bovis to the complex or genus level, the specificity and sensitivity of this technique were high, with detection limits of 0.1 fg of template DNA (11). The amplification of IS6110 and hsp65 targets was also specific and sensitive when tested with NTM strains, such as M. chelonae, M. malmoense, M. avium, M. fortuitum, M. marinum, and M. kansasii, or MTC members, such as M. bovis and M. africanum (27).
The convenience of this technique in routine settings processing a large collection of clinical material was demonstrated in our work. A method could be established with the initial application of multiplex PCR to all positive liquid cultures and the subsequent identification of NTM by PRA-PCR. The diversity of mycobacterial species found provided valuable clinical information concerning epidemiology and the clinical management of NTM infections in our institution.
No potential conflict of interest was reported.
Bensi EP contributed to laboratory work, preliminary analysis and discussion. Panunto PC contributed to laboratory work. Ramos MC contributed to laboratory work, sponsoring, planning, discussion and writing.