Craniosynostosis is a disorder characterized by the premature fusion of the calvarial sutures, causing an abnormal skull shape. Because this disorder occurs with a relatively high frequency, estimated at 1 in 2500 individuals, craniosynostosis represents a relevant medical problem (1). More than 100 syndromes have been shown to be associated with this disorder. It is believed that at least 20% of cases are due to single gene mutations or chromosome abnormalities (2). Although most cases can be considered both clinically and genetically heterogeneous, there is evidence that six genes are involved in many cases: MSX2, FGFR1, FGFR2, FGFR3, FBN1, and TWISTY (1).
The Msh homeobox 2 (Msx2) gene encodes a homeodomain transcription factor protein and is expressed in migrating cranial neural crest cells during development (3). Msx2 has central roles in craniofacial development (4) and limb and tissue formation (6). Furthermore, Msx2 overexpression was demonstrated to be associated with craniosynostosis in mice (5,7), indicating that normal craniofacial formation is dependent on the Msx2 dosage. Corroborating these findings, increases in the copy number in the MSX2 region have been reported in craniosynostosis patients (8-11).
In the present study, we describe the findings from a whole-genome array comparative genomic hybridization (aCGH) analysis of a four-year-old patient exhibiting craniosynostosis, microcephaly, psychomotor development delay, short stature, and cognitive impairment, among other abnormalities. We found a gain in a region of chromosome 5q35.2 that contains MSX2 and has been shown to be associated with craniosynostosis. Quantitative PCR confirmed the increase in the MSX2 copy number.
CASE DESCRIPTIONThis study was approved by the Hospital das Clínicas Institutional Ethics Committee, and written informed consent was obtained from the family. The propositus (a girl) was the first live born child of a healthy and non-consanguineous couple (the mother was 38 years old and the father was 40 years old at the time of conception). The mother is of Japanese descent, and the father is of African descent. The mother had experienced two previous miscarriages due to incontinence of the endocervical isthmus. The pregnancy was uneventful except for mild vaginal bleeding during the first trimester. The patient was born preterm (31 weeks) after vaginal delivery, with Apgar scores of 8 and 9, a weight of 1550 g, a length of 38.5 cm, and an OFC of 28.0 cm. At birth, the patient exhibited respiratory distress and underwent orotracheal intubation (3 days). The patient was hypotonic, had poor suction, and exhibited neonatal jaundice and metabolic disturbances (hyponatremia and hypocalcemia). She was discharged at 2 months of age at a weight of 1910 g. The patient also exhibited failure to thrive, recurrent otitis and global developmental delay. She was able to sit without support at 15 months and able to walk with no support and say simple words at 24 months.
Physical examination at 19 months showed a weight of 6500 g, a length of 68.5 cm, and an OFC of 42.5 cm. The patient's face was peculiar, with a narrowing of the frontal diameter, upslanting palpebral fissures, an enlarged nasal root, low and posteriorly rotated ears, bilateral convergent strabismus, a high arched palate and mild retrognathia. Neuroimaging studies (CT) revealed coronary craniosynostosis. No heart defects were observed. Abdominal ultrasonography showed a mild abnormality of the renal parenchyma, but the patient's kidney function is normal.
Phytohemagglutinin (PHA)-stimulated lymphocytes were subjected to G-banding karyotyping (500 bands) of the propositus and progenitors. The results were described according to the recommendations of the ISCN (2008). G-banding karyotype analysis of the patient showed a normal 46,XX pattern with no structural or numeric variations. The same was observed for both progenitors.
For the aCGH experiments, DNA from the patient and progenitors was extracted from blood samples using a QIAamp DNA Blood Midi kit (Qiagen, Germany). Labeling and hybridization reactions were performed as recommended by the manufacturer (Perkin Elmer, Norwalk). aCGH was performed with Constitutional Chip 4.0 (Perkin Elmer, Norwalk), which includes 5000 BAC clones spotted in duplicate with a resolution of at least 650 kb and human DNA segments of 100∼300 kb distributed through the whole genome.
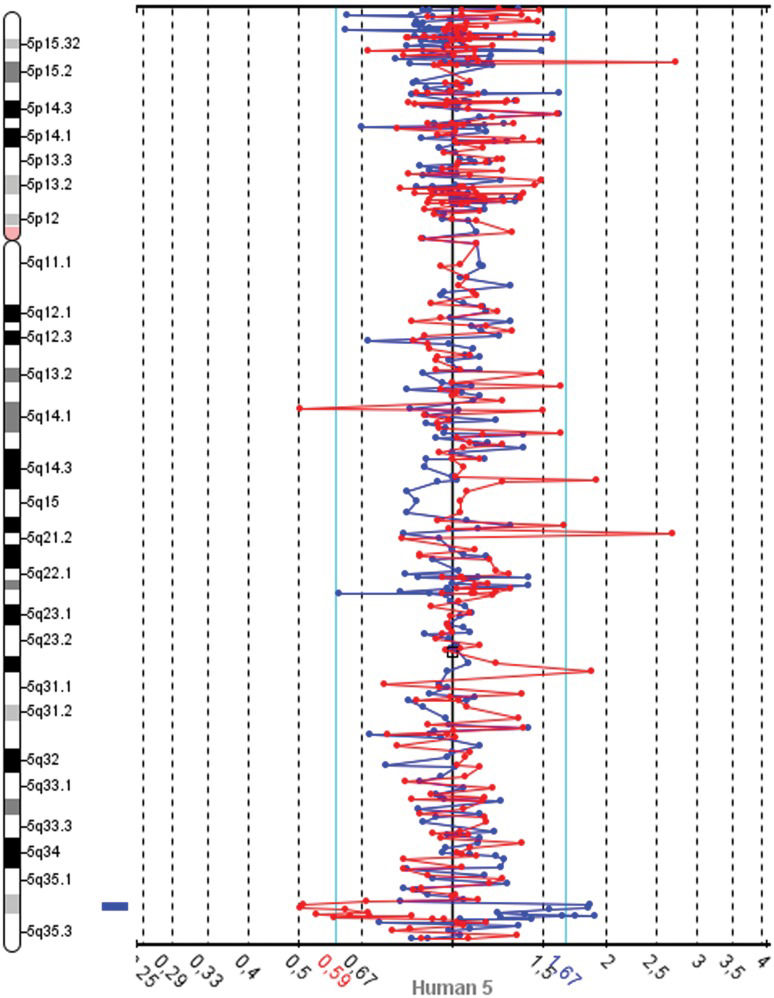
The slides were scanned with InnoScan 710 (Innopsys, Carbonne, France), and MAPIX 4.5 software was used to generate GPR files. Data analysis was conducted with SpectralWare® v2.3.3 aCGH Analysis System software (12) from Perkin Elmer. The computational parameters used were pin linear normalization, a threshold between 0.7 and 1.3, a Loess alpha of 0.1 for normalization and a confidence level of 95%. Median values were used for the interpretation of the results. The Database of Genomics Variants was consulted to determine the size of chromosome regions and to identify CNVs (13). These analyses revealed gains in two neighboring BAC clones on 5q32.2 and losses in 4q22.1, 8q24.3 and 9q34.2-34.3 (Table 1). A spectral image of chromosome 5 from the patient demonstrating the gain in 5q35.2 is presented in Figure 1. The patients revised karyotype was 46,XX.arr 4q22.1(88,282,368-88,336,595)x1, 5q35.2(173,984,926-174,145,340)(174,718,444-174,907,513)x5, 8q24.3(142,215,092-142,412,727)x1, 9q34.2q34.3(136,982,786-137,166,802)x1[hg18].
Clones that exhibited copy number variation detected through aCGH screening of the patient's genome.
| Clones | Status of alteration | Cytogenetic location | Genomic coordinates (GRCh36/ hg18) | Genes present in the region |
|---|---|---|---|---|
| RP11-203P12 | Loss | 4q22.1 | 4:88,282,368- 88,336,595 | KLHL8 |
| RP11-603O17 | Gain | 5q35.2 | 5:173,984,926- 174,145,340 | MSX2 |
| RP11-606P24 | Gain | 5q35.2 | 5:174,718,444- 174,907,513 | DRD1, SFXN1 |
| RP11-10J21 | Loss | 8q24.3 | 8:142,215,092- 142,412,727 | DENND3, SLC45A4 |
| RP11-153P4 | Loss | 9q34.2 | 9:135,531,566- 135,710,693 | SARDH, VAV2 |
| RP11-311J21 | Loss | 9q34.3 | 9:136,982,786- 137,166,802 | OLFM1 |
DNA samples from the patient and progenitors were used for MSX2 gene copy number assessment. A TaqMan copy number assay (Applied Biosystems, Foster City, CA) consisting of an MGB probe labeled with FAM and unlabeled PCR primers for MSX2 (assay Hs01821094_cn) was employed. Unlabeled primers and a VIC dye-labeled TAMRA probe for RNase P gene, which is known to be present in two copies in human diploid genome, were used for normalization. Reactions were performed as recommended by the manufacturer in triplicate. Duplex real-time polymerase chain reactions were conducted in a 7500 Real-Time PCR System, and the data were analyzed with CopyCaller™ Software v1.0, both from Applied Biosystems. Cycle thresholds (CT) were calculated using the relative quantification method (Applied Biosystems). Copy numbers were calculated by determining the difference in the CT between the target and control probes (ΔCT). Cycle thresholds greater than 32 were excluded from analyses. A quantitative PCR assay to detect MSX2 CNVs revealed two copies for both progenitors and five copies for the patient (Figure 2). Phenotypically normal subjects unrelated to the studied family also had two copies. The mean delta Ct, calculated as the FAM Ct divided by the VIC Ct, and the standard deviation of the delta Ct were 0.2192±0.13 for Subject 1, 0.5772±0.29 for Subject 2, 0.5807±0.1 for the patient's mother, -0.8858±0.12 for the patient, 0.3630±0.16 for Subject 3 and 0.7518±0.12 for the patient's father.
Assessment of MSX2 copy number variation. The estimated copy number variations for the patient, the progenitors, and three unrelated subjects are presented. The bars represent the variation in the calculated copy number. The values were ±0.41 for Subject 1, ±0.63 for Subject 2, ±0.23 for the patient's mother, ±0.79 for the patient, ±0.45 for Subject 3 and ±0.30 for the patient's father.
A search for similar abnormalities in DECIPHER (Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources) (14) returned three cases with chromosome 4 alterations related to that observed in the present study (cases 753, 994, and 249573). The phenotypes in these cases included mental retardation/developmental delay, short stature, atrial septum defects, hypotonia and eyelid defects, among other abnormalities. However, it is important to stress that no correlation between a loss in the 4q22.1 region and craniosynostosis has been described. In addition, no patient with case data reported in DECIPHER who had alterations in 8q24.3 or 9q34.2-34.3 similar to those in our patient exhibited craniosynostosis.
In a detailed study that investigated common CNVs in Asian population, it was found that the KLHL8 (4q22.1), VAV2 (9q34.2) and OLFM1 (9q34.3) genes exhibited copy number variations in phenotypically normal subjects (15), indicating that copy number losses in these genes most likely do not contribute to the craniosynostosis phenotype. Corroborating these findings, a search of the Online Mendelian Inheritance in Man (OMIM) database (16) returned no correlations between the genes KLHL8, VAV2, DRD1, SFXN1, DENND3, SLC45A4 and OLFM1 and clinical phenotypes. The SARDH gene, located in the 9q34.2 region, was shown to be associated with sarcosinemia, which is generally considered a benign condition unrelated to neurologic symptoms or significant clinical problems (17).
Among the CNVs found in the present work, the gain in the 5q35.2 chromosome region where the MSX2 gene is located has already been reported to be associated with craniosynostosis (8); however, the MSX2 gain was confirmed using molecular techniques in only four cases (8-11). MSX2 is a member of the homeobox MSX family and is related to Drosophila muscle segment homeobox (msh). MSX2 has important roles in tissue and organ development (3) and is also expressed in several regions of the developing skull. In mice, Msx2 is strongly expressed in osteogenic cells from calvarial sutures, and Msx2 activation is thought to initiate the release of factors from the dura mater that affect osteoblastic suture cells (18,19).
Consistent with this role of Msx2, transgenic mice that over expressed this gene exhibited premature suture closure and craniofacial abnormalities (5), whereas other evidence suggests that MSX2 haploinsufficiency leads to a delayed or incomplete closure of the opening between the frontal and parietal bones. MSX2 haploinsufficiency has been demonstrated to be responsible for the majority of foramina parietalia permagna (FPP) cases (20-22).
There are 39 reported cases of 5q distal trisomy, but craniosynostosis was observed in only ten of these cases. Because most craniosynostosis patients exhibit a distal 5q duplication, it is hypothesized that in patients with larger duplications, MSX2 expression may be inhibited due to extra copies of other genes (8). Additional cases may help to elucidate the validity of this hypothesis.
In conclusion, our analysis indicates that an increase in MSX2 copy number is correlated with this disorder, corroborating previous findings that subjects with gains in 5q35.2 in the MSX2 gene region exhibit craniosynostosis. In addition, the aCGH technique was shown to be useful for the detection of CNVs throughout the whole genome. Further studies should now be undertaken to determine which of these CNVs are normal individual variations and which are of clinical relevance, e.g., associated with specific syndromes.
We would like to thank the patient and her parents for their participation in our research. This study used data generated by the DECIPHER Consortium. A full list of the centers that contributed to the generation of the data is available from http://decipher.sanger.ac.uk and via e-mail from: decipher@sanger.ac.uk. We would specifically like to thank Dr. J. M. Friedman and Dr. McGillivray from the University of British Columbia for additional data on Decipher patient 994, Dr. Anne Philippe from Université René Descartes for data on Decipher patient 753 and Dr. Koen Devriendt for data on patient 249573. This study was supported by Fleury Group.
Pelegrino KO performed the aCGH experiments, the qPCR assays and the data analysis and interpretation and participated in the preparation and revision of the manuscript. Sugayama S was responsible for the patient examination and clinical description and participated in the preparation of the manuscript. Lezirovitz K performed the DNA extraction and participated in the aCGH experiments. Catelani AL performed the karyotype analysis. Kok F participated in the writing of the discussion section and in the revision of the manuscript. Chauffaille ML coordinated the study, designed the project, was responsible for fund obtaining, contributed to the data interpretation, manuscript writing and review.