The objective of this investigation was to identify the mechanism of decreased susceptibility to gentamicin in a Salmonella clinical isolate, leading to the detection of a aminoglycoside acetyltransferase gene found in a class 1 integron.
MethodsA multidrug-resistant Salmonella strain was recovered from feces of a traveler to Egypt. The antimicrobial susceptibility test to 12 antimicrobial agents was performed with the Kirby-Bauer method. The presence of class 1 integron was determined by PCR. The amplified product was recovered and sequenced in order to establish the genes carried. In addition, susceptibility to gentamicin C1a, gentamicin C1, sisomicin, neomycin, dibekacin, kanamycin, tobramycin, amikacin, netilmicin, apramycin, dactimicin, spectinomycin, streptomycin, lividomycin and butirosin, was established. The Champion™ pET101 Directional TOPO® Expression Kit was used to clone and express the aac(3)-I gene.
ResultsThe isolate was identified as Salmonella enterica serovar Haifa, showing resistance to nalidixic acid, tetracycline and decreased susceptibility to gentamicin. One integron with a size circa 1,500bp, encoding an aac(3)-Id plus aadA7 genes was observed. The analysis of the susceptibility to different aminoglycosides in the E. coli TOP10F’ transformed with the vector carrying aac(3)-Id gene showed resistance to gentamicin C1a, gentamicin C1, and dactimicin, in accordance with the presence of this enzyme but, was susceptible to sisomicin. The homology of the amino acid and nucleotide sequences with the AAC(3)-Id enzyme was of 100%.
ConclusionThe presence of the AAC(3)-Id enzyme was described for the first time in a S. Haifa.
el objetivo de este estudio fue identificar el mecanismo de sensibilidad disminuida a gentamicina en un aislamiento clínico de Salmonella, lo que nos condujo a la detección de un gen que codifica una acetiltransferasa modificante de aminoglucósidos localizada en un integron tipo 1.
Métodosla cepa multiresistente de Salmonella fue aislada de las heces de un viajero a Egipto. La susceptibilidad a 12 agentes antimicrobianos se determinó mediante Kirby-Bauer. La presencia de integron clase 1 se realizó mediante PCR. El producto de PCR amplificado del integrón fue recuperado y secuenciado para conocer los genes que contenía dicho integrón. Además se determinó la susceptibilidad a gentamicina C1a, gentamicina C1, sisomicina, neomicina, dibekacina, kanamicina, tobramicina, amikacina, netilmicina, apramicina, dactimicina, espectinomicina, estreptomicina, lividomicina y butirosina. El kit de expresión Champion™ pET101 Directional TOPO® fue utilizado para clonar y expresar el gen aac(3)-I.
Resultadosel aislamiento fue identificado como Salmonella enterica serovariedad Haifa, el cual presentaba resistencia al ácido nalidixico, tetraciclina y sensibilidad disminuida a gentamicina. Se observó la presencia de un integron tipo 1 con un tamaño de 1,500bp en el que se encontraron dos genes (aac(3)-Id y aadA7). El análisis de la sensibilidad a diferentes aminoglucósidos de la cepa de E. coli TOP10F’ transformada con el vector que contenia el gen aac(3)-Id demostró resistencia a gentamicina C1a, gentamicina C1, y dactimicina, en concordancia con la presencia del enzima pero era susceptible a sisomicina. La secuencia de aminoácidos presentaba un 100% de identidad con el enzima AAC(3)-Id.
Conclusiónla presencia del enzima AAC(3)-Id ha sido descrita por primera vez en S. Haifa.
Traveler's diarrhea (TD) is a frequent health problem among travelers to developing countries. This illness may be due to a large variety of microorganisms, among these, Salmonella is one of the most frequent following diarrheogenic Escherichia coli and Shigella spp.3,21. Diarrhea associated with Salmonella spp. is usually self-limited and does not require antibiotic therapy. However, in specific cases, due to both the severity or the duration of the symptoms, antibiotic treatment is required. Unfortunately, antimicrobial resistance levels among diarrheagenic pathogens have increased in recent years, and Salmonella spp. is not an exception3,21.
Acquisition of resistance may be related to two different mechanisms: 1. Transferable, such as plasmids or transposons, and 2. Non transferable, usually associated with chromosomal point mutations7,19. The gastrointestinal environment serves as a reservoir for integron-bearing strains and since integrons are carried on plasmids and transposons, antibiotic selective pressure can potentate the dissemination of antibiotic resistance genes through these genetic elements15.
To date, nine classes of integrons have been described16. Of these, the most relevant at a clinical level are those belonging to classes 1 and 2. The integrons of these two aforementioned classes usually carry gene-cassettes encoding for antibiotic resistance mechanisms. Among these gene-cassettes, the aminoglycoside-modifying encoding genes are considered the most prevalent6. The aim of this work was to investigate the mechanism of decreased susceptibility to gentamicin in a clinical isolate of Salmonella enterica serotype Haifa.
MethodsBacterial isolateA Salmonella isolate recovered from feces of a traveler with diarrhea was identified by different typing methods, including biochemical tests and serotyping using somatic and flagella antiserum11.
Antimicrobial susceptibilityA preliminary antimicrobial susceptibility test was performed, using an agar diffusion method with commercially available disks (Becton Dickinson) to the following antibiotics: ampicillin, amoxicillin plus clavulanic acid, nalidixic acid, tetracycline, trimethoprim/sulphametoxazole, chloramphenicol, gentamicin, amikacin, imipenem, norfloxacin, ciprofloxacin and ceftazidime. Interpretation of results was performed according to the Clinical Laboratory Standards Institute (CLSI) guidelines. E. coli ATCC 25922, E. coli ATCC 35218 and Pseudomonas aeruginosa ATCC 27853 were used as controls4.
To assess an activity pattern, susceptibility to gentamicin C1a, gentamicin C1, sisomicin, neomycin, dibekacin, kanamycin, tobramycin, amikacin, netilmicin, apramycin, dactimicin, spectinomycin, streptomycin, lividomycin and butirosin, the disk diffusion method on Mueller-Hinton agar was used. These disks were manually prepared adding 30μg of each antibiotic to 10mm sterile blank filter disks. E. coli ATCC 25922 was used as a susceptible control strain. Reductions of the inhibition zone were considered as the result of aminoglycoside-modifing enzyme (AME) activity. Antimicrobial susceptibility levels of gentamicina were also established by the E-Test method following the manufacturer's instructions.
Detection of class 1 integronsThe presence of class 1 integrons was determined by PCR using the primers and conditions previously described12. The amplified products were gel recovered using the Wizard SV Gel and PCR Clean-up System Kit (Promega, Madison, USA) and sequenced using the BigDye Terminator v3.1 Cycle Sequencing Kit (Perkin Elmer, Emeryville, USA).
Plasmid analysisPlasmid DNA was isolated as described by Kado and Liu8. The plasmid extracted DNA was resolved by electrophoresis on 0.8% agarose gel and stained with ethidium bromide (0.5mg/L).
ConjugationBacterial conjugation experiments were performed using E. coli J53 (F-, pro, Gm S, Rif R, Lac +) as the receptor strain as previously described12, and were repeated three times.
DNA amplification and cloning of the aac(3)-I geneThe Champion™ pET101 Directional TOPO® Expression Kit (Invitrogen, USA) was used to clone and express the aac(3)-I gene, following the manufacturer guidelines. Briefly, the entire AAC(3)I encoding gene was amplified using the forward primer AAC3IF: 5′-CAC CGT GTC AGT CGA AAT CAT C-3′ and the reverse primer AAC3IR: 5′-GGC ATG ATT TTT ACT CTG C-3′. The amplification product was resolved by electrophoresis on a 2% agarose gel stained with ethidium bromide.
The PCR product was gel recovered, using a Wizard SV Gel and PCR Clean-Up System (Promega, Madison, Wi) and was directly cloned into the pET vector and transformed into E. coli TOP10F’. Transformed E. coli strain were spread on a selective plate with ampicillin and incubated overnight at 37°C. Plasmids were isolated from several colonies and then analyzed by PCR using the AAC3IF primer and the specific vector primer T7 Reverse. The isolated plasmid was used to transform E. coli BL21 Star (DE3) for expression studies. The cloned insert was expressed in plates containing IPTG (1mM).
Results and discussionA Salmonella enterica serovar Haifa was isolated from feces of a traveler with diarrhea returning from Egypt. This strain showed resistance to nalidixic acid, tetracycline and decreased suscep-tibility to gentamicin, while remaining susceptible to ampicillin, amoxicillin plus clavulanic acid, ceftazidime, cotrimoxazole, chloramphenicol, amikacin, imipenem, norfloxacin, and ciproflo-xacin.
The isolate was investigated for the presence of class 1 integrons. One amplicon of circa 1500bp was detected. The sequence of this amplicon revealed the association of the integron with aac(3)-Id plus ant(3″) (also named aadA7) aminoglycoside-resistance genes (Figure 1). The detected aac(3)-Id nucleotide sequence showed amino acid and nucleotide homologies of 100% both with the aac(3)-Id and aac(3)-Ie genes located in similar integrons in Salmonella enterica serovars Newport and Kentucky5,9, as well as in Vibrio fluvialis1 (GeneBank access: AY458224, AY463797 and AB114632). Meanwhile, the homology with aac(3)-Ia , aac(3)-Ib and aac(3)-Ic was lower. The ant(3″) did not show differences with other nucleotide sequences previously reported.
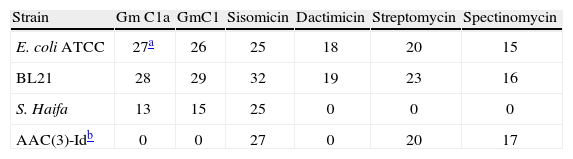
The Salmonella enterica serovar Haifa isolate showed a resistance pattern partially consistent with the presence of an AAC(3)-Id plus an ANT(3″) aminoglycoside nucleotidyltransferase, with resistance or decreased susceptibility to gentamicin C1a, gentamicin C1, dactimicin, streptomycin and spectinomycin, but susceptible to sisomicin an aminoglycoside also considered a substrate of the AAC(3)I-type enzymes. (Table 1)10. In order to establish the exact role of the aac(3)-Id in the aminoglycoside resistance pattern detected, the gene was cloned in an expression vector. In the presence of IPTG, the transforming strain showed resistance to gentamicin C1a, gentamicin C1 and dactimicin, but lost the resistance to streptomycin, spectinomycin (Table 1), remaining susceptible to sisomicin. When the MIC of gentamicin was established in this transforming strain the results shows that possess a MIC of 48μg/ml irrespective of the presence or absence of IPTG.
Aminoglycoside modifying enzymes are commonly located within integrons in pathogens causing TD, such as Shigella spp.13. In fact, different reports both in pathogens causing TD or not, considered that cassettes encoding for these genes are the most frequently found among integrons2. To date, 5 different aac(3)-I encoding genes have been described in the literature: aac(3)-Ia22, aac(3)-Ib17, and aac(3)-Ic14, aac(3)-Id (1) and aac(3)-Ie9. However, out of them, the aac(3)-Id and the aac(3)-Ie genes, showed the same nucleotide sequence. As in the above mentioned aac(3)-I-like genes, the aac(3)-Id described in the present study is located within an integron, together with the ant(3″) gene. Analysis of the transformed E. coli strain clearly showed that the aac(3)-Id gene product is responsible for the resistance to gentamicin C1a, gentamicin C1, and dactimicin, while resistance to streptomycin and spectinomycin was probably associated with the detected ant(3″) gene20,23.
The genetic location of the genes encoding the different acetyltransferases varies widely; for instance, the aac(2′)-I gene is almost exclusively located in the chromosome in Providencia spp. and Proteus spp.18, whereas most of the genes of the AAC(6) family are present in plasmids18. To establish the genetic location of the aac(3)-I gene, both plasmid analysis and conjugation experiments were performed, showing negative results. However, the PCR of the aac(3)-Id using chromosomal DNA extracted from an agarose gel as a DNA template was positive. The amplification of the gyrA gene was used as a control (data not shown). Therefore, our results suggest the chromosomal location of the integron.
The phenotypic characteristics of the strains analyzed (decreased susceptibility to gentamicin and susceptibility to sisomicin) suggested the presence of a new aminoglycoside acetyltransferase gene. Nevertheless, the genetic study show the presence of AAC(3)-Id enzyme, before described in Salmonella enterica serovar Newport. The phenotype (decreased susceptibility to gentamicin and susceptibility to sisomicin) shown by the strain may be explained by a posttranslational change in the conformation of the enzyme. However, structural studies would be needed to show this hypothesis.
A Salmonella enterica serovar Haifa carrying a aac(3)-Id gene was identified in a class 1 integron for the first time. This result shows the potential of integrons to carry and spread resistance genes.
This study was funded by Ministerio de Sanidad y Consumo, Instituto de Salud Carlos III – FEDER, Spanish Network for the Research in Infectious Diseases (REIPI RD06/0008), FIS 04/0068, and grant 2005 SGR 0444 from the Departament d’ Universitats, Recerca i Societat de la informació de la Generalitat de Catalunya, Spain to J.V. and by the DGA/Group of Ecology of Bacterial resistance, Spain. R.C. has a fellowship from Fundación Carolina and BBVA, Spain. J.R. research is supported by project CP05/0130.