The human body is populated by myriads of microorganisms throughout its surface and in the cavities connected to the outside. The microbial colonisers of the intestine (microbiota) are a functional and non-expendable part of the human organism: they provide genes (microbiome) and additional functions to the resources of our species and participate in multiple physiological processes (somatic development, nutrition, immunity, etc.). Some chronic non-communicable diseases of developed society (atopias, metabolic syndrome, inflammatory diseases, cancer and some behaviour disorders) are associated with dysbiosis: loss of species richness in the intestinal microbiota and deviation from the ancestral microbial environment. Changes in the vertical transmission of the microbiome, the use of antiseptics and antibiotics, and dietary habits in industrialised society appear to be at the origin of dysbiosis. Generating and maintaining diversity in the microbiota is a new clinical target for health promotion and disease prevention.
El cuerpo humano está poblado por miríadas de microorganismos en toda su superficie y en las cavidades conectadas con el exterior. Los colonizadores microbianos del intestino (microbiota) son parte funcional y no prescindible del organismo humano: aportan genes (microbioma) y funciones adicionales a los recursos de nuestra especie y participan en múltiples procesos fisiológicos (desarrollo somático, nutrición, inmunidad, etc.). Algunas enfermedades crónicas no transmisibles de la sociedad desarrollada (atopias, síndrome metabólico, enfermedades inflamatorias, cáncer y algunos trastornos de la conducta) se asocian a disbiosis: pérdida de riqueza de especies en la microbiota intestinal y desviación del entorno microbiano ancestral. Los cambios en la transmisión vertical del microbioma, el uso de antisépticos y antibióticos, y los hábitos dietéticos de la sociedad industrializada parecen estar en el origen de la disbiosis. Generar y mantener diversidad en la microbiota es un nuevo objetivo clínico para la promoción de salud y prevención de enfermedades.
A person's health depends on their biology (genetics, development, ageing), lifestyle (diet, exercise, medications taken, substance abuse, etc.), environment (physical, chemical, biological, psychosocial, sociocultural external factors, etc.) and their health system (use of services, efficacy, efficiency, etc.). Microbial colonisation plays a role in many of these variables in one way or another. The experimental and clinical research of the last decade has revealed the functional impact of the microbial communities that live in the intestines of animals, including humans. The intestinal or gut microbiota plays a key role in health and disease. Although the optimal symbiosis between microorganisms and host is not yet fully understood, and the most appropriate proportion or combination of microorganisms for each individual is not known, there is nevertheless some basic scientific evidence that doctors should be aware of and apply in clinical practice.
Composition and functions of the gut microbiotaMicrobial colonisation and the development of the gut microbiota proper begins at birth, even though foetal exposure to microorganisms may be limited.1 The gut microbiota plays a key role in the development of an individual's immune system and homeostasis,2 and the initial colonisation phases are crucial. Germ-free animal experiments have shown that microbial colonisation in early life induces trophic and immune functions, which does not occur if colonisation occurs in adulthood.3,4
Microbiota acquisition is influenced by several factors5: type of birth, gestational age, initial diet, exposure to antibiotics, etc. Babies born vaginally have an initial microbiota similar to that of the maternal vagina, while babies born by caesarean section have microbiota similar to the skin or the environment.6 Babies born prematurely have low levels of anaerobes, such as Bifidobacterium or Bacteroides, and higher levels of Enterobacteriaceae, which include potential pathogens (Escherichia coli or Klebsiella pneumoniae).7 Antibiotics, including any administered prophylactically to the mother, affect gut microbiota acquisition.8
The microbiota of children who were exclusively breastfed has been found to contain more beneficial microorganisms, such as bifidobacteria, than children given formula milk.9 Other factors, such as older siblings, pets and a rural versus urban environment also have an impact.5 The introduction of solid food in place of breast milk triggers significant changes, and the Bacteroidetes and Firmicutes phyla become dominant for the rest of the child's life. Microbial diversity gradually increases, as does the capacity to break down complex carbohydrates and xenobiotics and to produce vitamins. The microbiota of a three-year-old already resembles that of an adult,5 although some microbial groups are only complete in adolescence.10
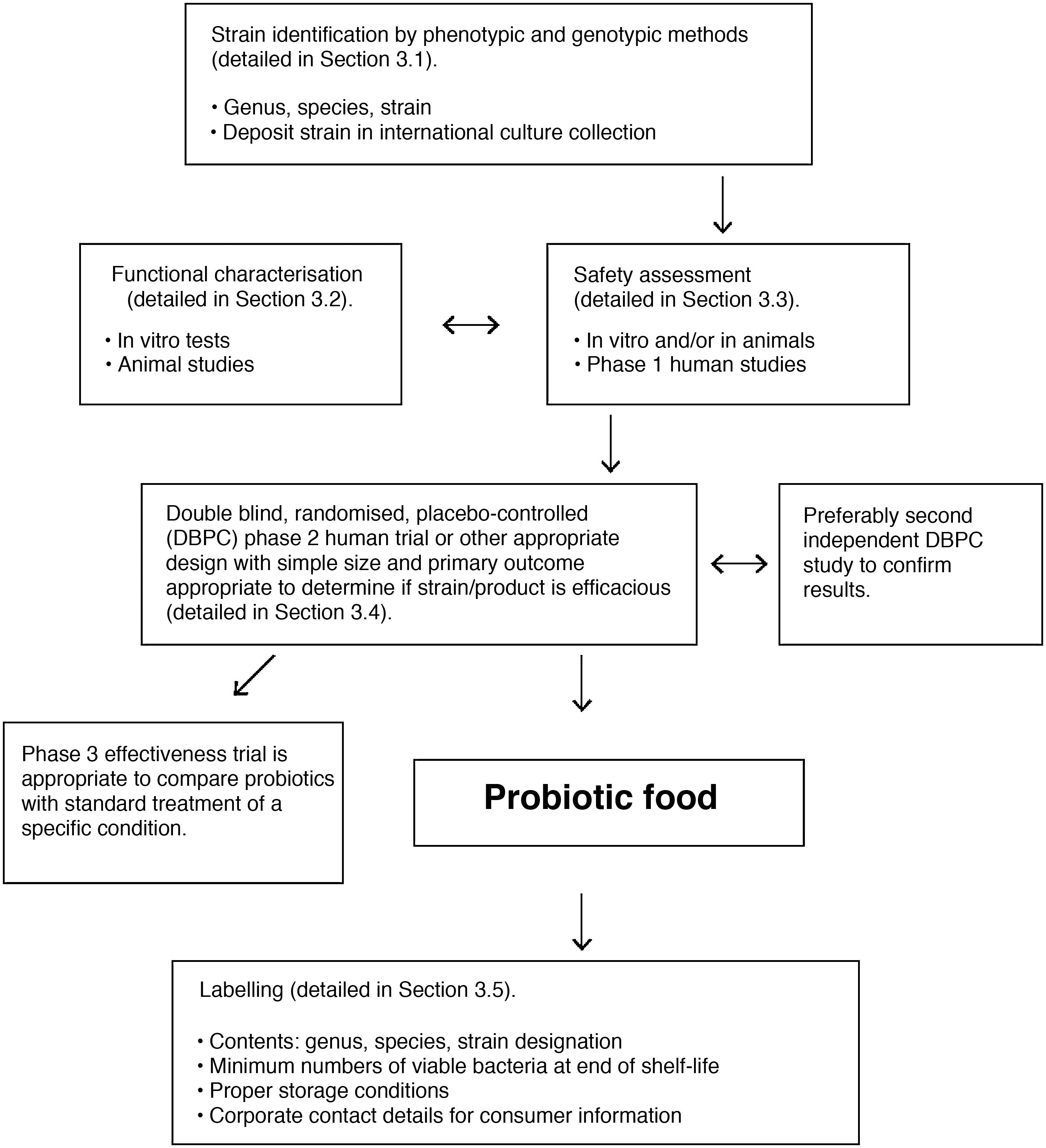
In adults, 90% of the gut bacteria belong to two phyla: Bacteroidetes and Firmicutes. The remaining 10% is made up of Proteobacteria, Actinobacteria, Fusobacteria and Verrucomicrobia, as well as a few species from the Archaea domain.11 The human gut microbiota also includes yeasts, phages and protists. The viral component is dominated by bacteriophages. These are known to play a crucial role in the make-up of the ecosystem by controlling the proliferation of dominant species and horizontal gene transfer, but the vast majority of viral sequences share little or no homology with reference databases.12 Yeasts form a relatively non-diverse community, with fewer than 20 species having been identified in a healthy adult intestine. Their relative abundance is four to five orders of magnitude less than that of bacteria, but their cell size and genome are much larger, and they contribute functional resources that are integrated into the ecosystem.13
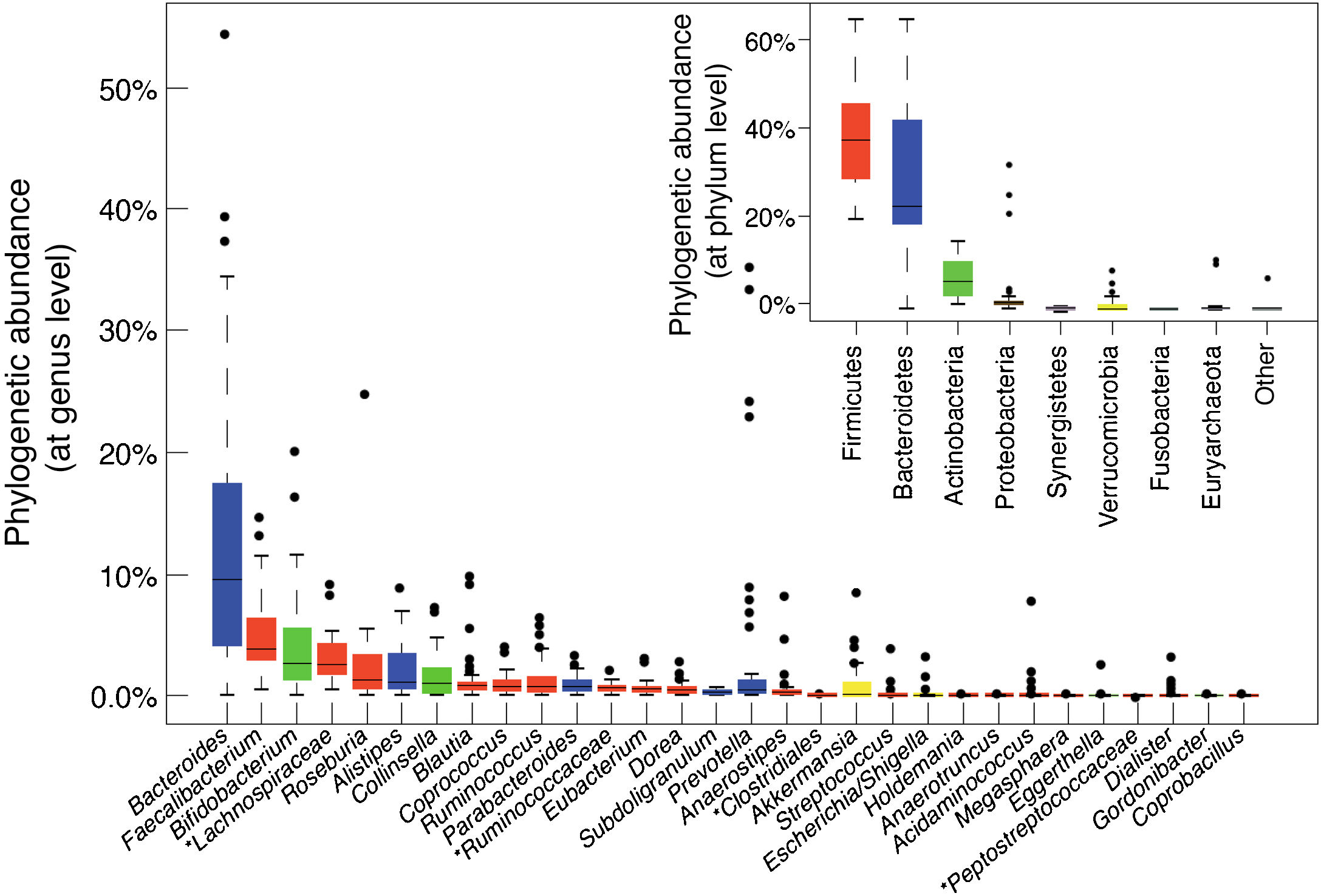
The main bacterial genera are shown in Fig. 1. In terms of strain, each individual hosts a distinctive pattern of microbial communities containing many unique strains that are not found in other individuals. Although there are differences between the various sections of the digestive tract, as well as between the faeces and intestinal mucosa of a single individual, the same strains are generally detected but in different proportions. Longitudinal studies have shown that factors such as diet, medication, travel and transit time through the colon affect the microbial composition of the faecal samples of a single individual, but differences between individual subjects are much greater than intra-individual variations. Although these fluctuations may be significant, the microbial ecosystem tends to revert to its typical pattern. Resilience is an important characteristic of a healthy gut microbiota ecosystem. It is characterised by the ability to restore the gut microbiota balance following a disturbance, such as an episode of acute diarrhoea or antibiotic treatment.14
Bacterial composition of the gut microbiota by genus and phylum or class, obtained by next-generation sequencing of DNA extracted from the stool samples of Spanish and Danish subjects of the MetaHIT cohort (data published by Arumugam et al.11).
Although the microbial composition of each individual is unique, the overall structure forms patterns that are repeated in different individuals, defined as enterotypes. The enterotype concept suggests that the microbial ecosystem in the human intestine relies on internal symbiotic relationships between the different members of the microbial community, probably determined by the metabolic or social networks in which they are integrated. These interactions explain the stability and resilience of a fluctuating ecosystem. Enterotype 1 individuals have an abundance of Bacteroides, enterotype 2 an abundance of Prevotella and enterotype 3 an abundance of Ruminococcus or Bifidobacterium.11 Diet is one of the main determinants of enterotype (section 5.1).
The onset of senescence marks the start of a new period of instability. Ageing is associated with a loss of microbial diversity and changing levels of some microorganisms.15,16 Changes are associated with an immune dysfunction known as "inflammaging", characterised by increased inflammation and an inhibited capacity to generate adaptive immune responses, as well as falling levels of microorganisms with anti-inflammatory properties, such as Faecalibacterium prausnitzii, and of other beneficial bacteria such as bifidobacteria.17 Modulating the gut microbiota or administering some of these microorganisms could help to curb age-related physiological decline. In fact, administering Akkermansia muciniphila can reduce degenerative symptoms and extend life expectancy in a progeria model.18
Digestion and metabolismThe gut microbiota plays a key role in the host's digestion and metabolic regulation. Part of the food we eat is not broken down completely by human enzymes. Residues that are not absorbed end up in the colon, inhabited by a high density of microorganisms with additional metabolic resources. The most common process is the fermentation of complex carbohydrates, which generates short-chain fatty acids (SCFAs), primarily acetic acid, propionic acid and butyric acid. These are used by enterocytes as a source of energy or end up in the bloodstream and are transported to distal organs, where they perform important functions.19 The production of bioactive compounds like group B vitamins or vitamin K is also important.20,21
The gut microbiota transforms inactive dietary compounds into bioactive molecules. An example of this is the transformation of some soy isoflavones into compounds with oestrogenic activity, such as equol.22 The microbiota can also generate potentially toxic compounds, such as the formation of trimethylamine from dietary choline and carnitine.23 Trimethylamine, which is absorbed by the liver and transformed into trimethylamine oxide (TMAO), is a cardiovascular disease risk factor.24
Microbial energy extraction from food is variable and depends on the microbial composition. Because the microbiota also regulates lipid storage, its role in obesity and metabolic syndrome is also being investigated.25 Faecal transplant from obese rats to rats of normal weight can transfer the phenotype, causing the latter to gain weight and store more fat.26 This shows that the gut microbiota performs a significant regulatory role in an individual's metabolic homeostasis.
Maturation and regulation of the barrier function and immune systemThe gastrointestinal tract (GI tract) has developed defence mechanisms against adverse environmental agents to which it is exposed orally (allergens, contaminants, pathogens, etc.), while at the same time tolerating resident commensal microbiota and dietary proteins. The gut microbiota affects the development and function of the immune system. As such, any disruption to this balance with the host can give rise to immune dysregulation and contribute to the onset of chronic inflammatory and autoimmune diseases.27
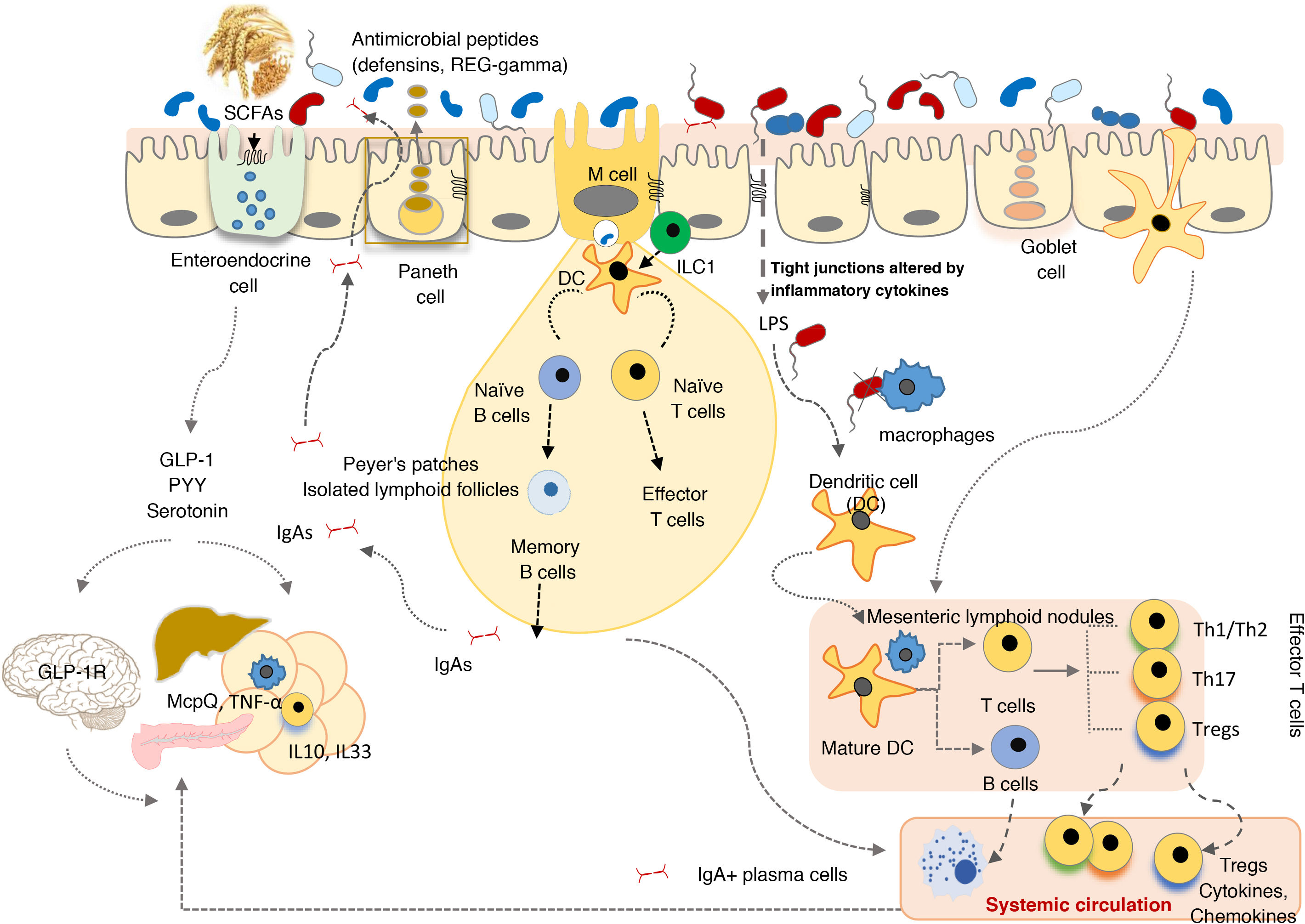
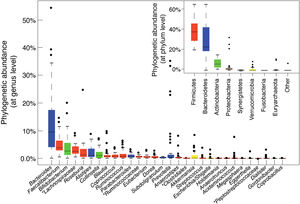
The GI tract is a selective barrier made up of epithelial cells. These hinder both resident and temporary microorganisms from coming into direct contact with specialised immune cells of the lamina propria, and their systemic propagation, thereby contributing to immunological homeostasis (Fig. 2). This barrier consists of enterocytes (90–95%), enteroendocrine cells, goblet cells, M cells, which are involved in antigen uptake, and Paneth cells. Goblet cells secrete mucin glycoproteins, which are assembled in the large intestine to generate two layers of mucus. The outermost layer traps a large number of microbes, preventing them from getting to the epithelium and facilitating their elimination in faeces.28 The small intestine is inhabited by Paneth cells responsible for the secretion of antimicrobial peptides (defensins, REG-gamma, etc.). These inhibit the growth of certain bacteria and prevent them from coming into direct contact with the epithelium.29 In turn, commensal bacteria regulate the expression of mucin-encoding genes (MUC-2, MUC-3) and modify their glycosylation pattern, as well as the production of antimicrobial peptides, which helps to regulate adhesion, colonisation and microbial invasion.27,29
The commensal microbiota also exerts a trophic effect, influencing the proliferation of epithelial cells and maintaining tight intercellular junctions. This helps to enhance the role of the epithelium as a physical barrier against the entry of exogenous agents.30 The production of secretory immunoglobulin A (IgAs) is another defensive mechanism that hinders bacterial access to the mucosa. Dendritic cells recognise and capture small amounts of bacteria and interact with the B and T cells in Peyer's patches, activating the production of specific IgAs. These IgAs are transported through the epithelium and, once in the lumen, bind to gut bacteria, where they help to control pathogens, neutralise toxins and promote their elimination.31
In the lamina propria, macrophages engulf and eliminate any microorganisms that may have penetrated the intestinal epithelium. Dendritic cells appear to be more involved in the coordination between innate and adaptive immunity. The bacteria or other antigens captured by dendritic cells are carried to the mesenteric lymph nodes, where they may be involved in T cell differentiation into effector or regulatory cells, depending on the type of antigenic stimulus and inflammatory tone. Intestinal homeostasis is maintained by a system of controls and balances between inflammatory effector T cells, which include Th1 cells (CD4+ and CD8+ that migrate to the epithelium and become intraepithelial lymphocytes), Th17 cells and Foxp3+ anti-inflammatory regulatory T cells (Tregs) that participate in the development of tolerance. Commensal bacteria are involved in the development of Treg cells, the irresponsiveness of effector T cells and the induction of T cell apoptosis or anergy, which prevents chronic inflammation and autoimmune phenomena. Moreover, activated dendritic cells interact with the B and T cells in Peyer's patches, inducing the production of both secretory IgAs and IgAs that provide systemic protection.27
Innate lymphoid cells (ILCs) are found in the intestinal epithelium and lamina propria32 and are involved in maintaining appropriate immune responses to different microorganisms, enhancing adaptive immunity and regulating intestinal mucosa tissue inflammation and repair. ILC functions are also regulated by the commensal microbiota.
The microbiota and cells of the innate immune system interact through microbial molecular pattern recognition receptors (toll-like receptors [TLRs], NOD-like receptors [NLRs], inflammasomes, etc.) or through metabolites (tryptophan, indoles, butyrate) produced by the microbiota (aryl hydrocarbon receptor [AhR], G protein-coupled receptors [GPCRs]). These stimuli activate both the aforementioned barrier functions as well as the synthesis of other mediators (cytokines, co-stimulatory molecules, etc.) that regulate the response of specialised immune cells in the gut-associated lymphoid tissue, as well as coordinate its actions to prevent pathogenic invasion and promote the development and tolerance of harmless antigens. For example, TLR2 activation by lipoteichoic acids of Gram-positive bacteria regulates tight junctions between enterocytes and helps prevent paracellular permeability from increasing, which could give rise to excessive immune activation and systemic inflammation. Clostridium clusters IV and XIVa induce regulatory T cell expansion in the lamina propria as well as systemic expansion. This is believed to be through the production of SCFAs, which act on G protein-coupled receptors and modulate immune cell function.33
Influence on the neuroendocrine systemThe microbiota and metabolites that are generated in the intestine from the diet emit neural and endocrine signals that affect distant organs and tissues. This is how the microbiota plays a role in functions as diverse as the regulation of energy balance (intake, energy expenditure, glucose metabolism, etc.), as well as other functions that depend on the nervous system, including cognitive, mood and behavioural functions (microbiota-gut-brain axis).
By generating SCFAs, the commensal microbiota exerts trophic effects on the intestinal mucosa and activates various receptors (GPR41 and GPR43). These stimulate the production of enteroendocrine hormones by L cells, such as glucagon-like peptide (GLP-1) or peptide tyrosine-tyrosine (PYY). SCFAs and enteroendocrine peptides help to regulate energy homeostasis by modulating aspects such as glucose metabolism, insulin sensitivity, thermogenesis and appetite. This occurs through endocrine effects on peripheral organs (liver and white and brown adipose tissue), as well as by neuronal signalling pathways (enteric and autonomic nervous system) that reach the central nervous system, where the signals that regulate energy homeostasis in both the short term (GLP-1) and the long term (insulin, leptin) are integrated.34 It has more recently been suggested that other metabolites deriving from the metabolic activity of the microbiota on diet, such as indoles, may increase the sensitivity of colonic afferent neurons to GLP-1, bolstering the effects of this hormone on energy homeostasis.34 Moreover, the gut microbiota is involved in the metabolism and enterohepatic circulation of bile salts and helps to form secondary bile acids. These interact with the TGR5 receptor, inducing differentiation and increasing the number of L cells and GLP-1 secretion, which also has a beneficial effect on oral glucose tolerance.35
Epithelial cells also play a role in the synthesis of glucocorticoids by performing an anti-inflammatory and endocrine function, and both glucocorticoid synthesis as well as the expression of its receptors can be affected by the composition of the gut microbiota.36 Commensal bacteria are involved in the circadian regulation of glucocorticoids in a manner that is dependent on the hypothalamic-pituitary-adrenal axis. Gut microbiota depletion impairs the expression of those genes involved in rhythms of corticosterone production in the ileum, which gives rise to significant alternations, such as the sustained systemic hypersecretion of corticosterone and, in turn, a state of hyperglycaemia, insulin resistance and hypertriglyceridaemia.37
The microbiota is also directly or indirectly involved in the synthesis of neuroactive compounds, including several neurotransmitters (serotonin, dopamine, gamma-aminobutyric acid [GABA], etc.) that affect brain functions, behaviour, metabolism and immunity. The microbiota’s involvement in serotonin synthesis seems to be particularly significant given that up to 90% of this neurotransmitter, which performs key central and peripheral functions, is synthesised in the intestine. In the central nervous system, serotonin plays a key role in regulating mood, appetite and cognitive functions, while in the intestines it regulates inflammation and motility.38 The gut microbiota may be involved both in reducing the levels of serotonin (due to its metabolising action on tryptophan that acts as a precursor), and in its production, by stimulating the expression of the host genes (tryptophan hydroxylase 1) involved in its synthesis, potentially through the stimulating effect of SCFAs.39 Dysregulation of the serotonergic system is also associated with chronic inflammatory diseases and diet-induced obesity. The microbial modulation of serotonin biosynthesis and of the expression of its receptors mitigates intestinal inflammation and depressive symptoms.38
Various gut bacteria encode tyrosinases capable of transforming tyrosine into l-dihydroxyphenylalanine (L-DOPA), which in turn gives rise to the synthesis of catecholamines, such as dopamine, norepinephrine and epinephrine. Dopamine plays an important role in the reward system, which helps to regulate eating behaviour as well as mood. However, the microbiota of some individuals can break down the drug l-DOPA, reducing its bioavailability and its efficacy in treating Parkinson's disease.40
GABA is produced by several gut bacteria thanks to the enzyme glutamate decarboxylase, which catalyses the decarboxylation of glutamate. This enzyme is involved in tolerance to acidic pH and in maintaining bacterial intracellular homeostasis. GABA is an important neurotransmitter with inhibitory effects on the brain, and its dysfunction is associated with disorders such as autism and schizophrenia. Preclinical studies suggest that the administration of certain probiotic and prebiotic bacteria may increase concentrations of GABA or its receptors in the brain, which in turn may have a positive impact on these disorders.41
Eubiosis and dysbiosisStable microbial communities that reside in niches live in a state of equilibrium and typically comprise an abundance of species that have a commensal and mutualistic relationship with the host, such that both the host and the microbiota benefit from the symbiosis. This is known as "eubiosis". In contrast, the term "dysbiosis" describes an imbalance that disturbs the state of symbiosis. It is recognised by qualitative and/or quantitative changes in microbiota composition and functions. However, determining what a "normal" microbiota is, both in terms of its composition and its functions, is not easy due to the multiple factors that influence its configuration, as mentioned earlier, and due to the significant inter- and intra-individual variability in physiological conditions.42
States of dysbiosis typically involve the loss or insufficient numbers of beneficial species that are usually dominant, and the increased abundance of minority species that often include pathobionts or opportunistic pathogens. Changes can be specific to each niche and each disease, and can alter the overall structure of the microbiota, or give rise to the loss or acquisition of particular species. For example, inflammatory bowel disease usually involves the loss of butyrate-producing bacteria (Faecalibacterium, Roseburia, Eubacterium)43 which, in the case of antibiotic-associated diarrhoea, coincides with an overgrowth of opportunistic species, such as Clostridioides difficile in pseudomembranous colitis.44 In colorectal cancer, the increased relative abundance in the faeces of Fusobacterium, a genus of oral microbiota, is common.45
Multiple factors may contribute to the onset of dysbiosis, including the use of antibiotics and other medicines, stress, genetic factors, diet, lifestyle, etc. If the triggering factor is intense and long-lasting, the process can lead to disease, typically chronic or recurrent and inflammatory. In early life, maternal dysbiosis can lead to altered vertical transfer and affect the initial acquisition of gut microbiota, with potential short- and long-term consequences.5
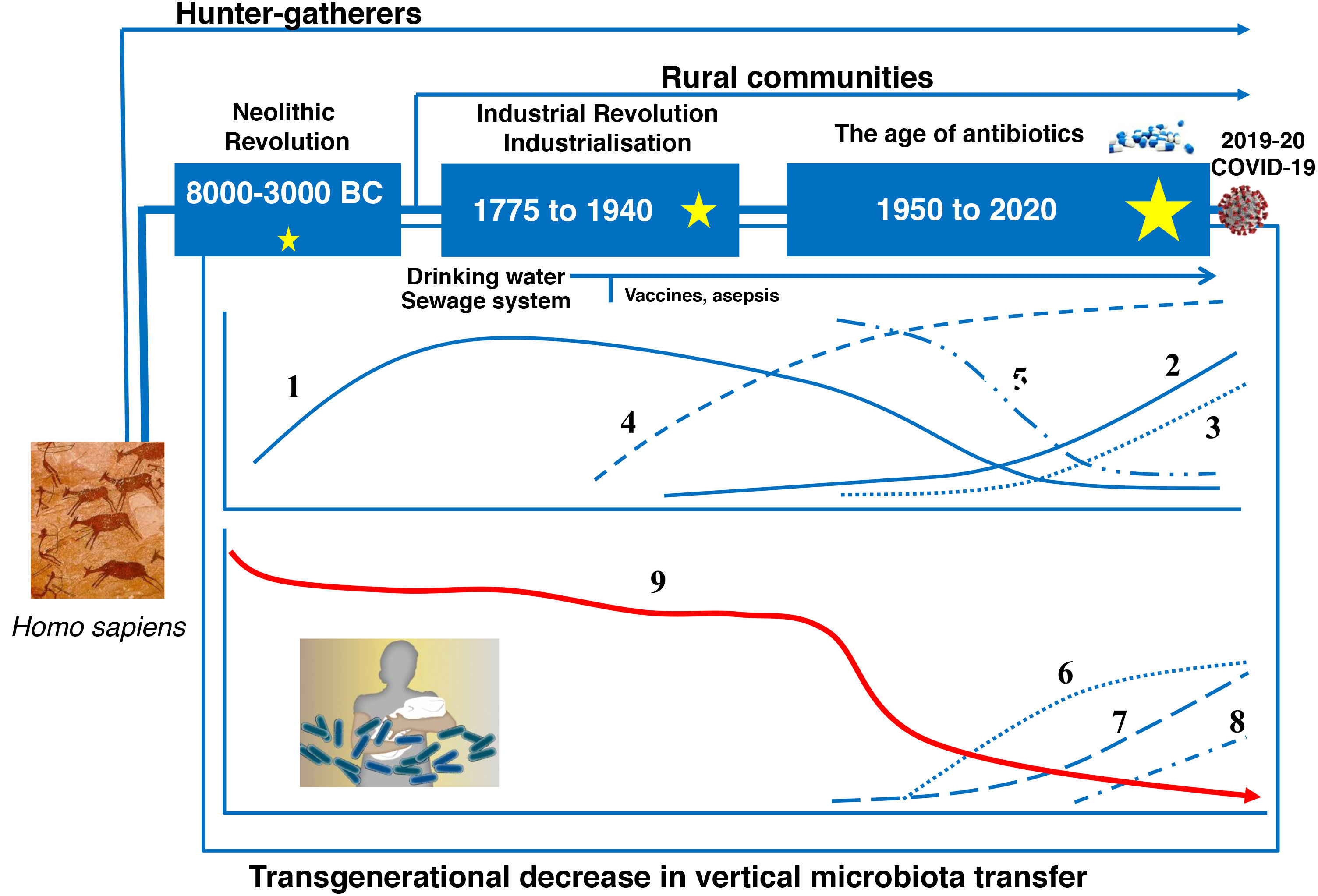
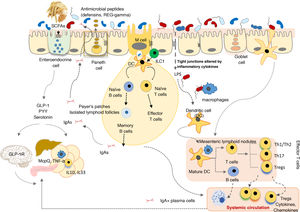
Evolution of the human gut microbiotaRecent advances in microbial ecology and genomics show that major disturbances in human microbial populations are closely correlated to the major stages of the Anthropocene: a) the "Paleoanthropocene" that began with the emergence of agriculture and livestock in the Neolithic period; b) the industrial revolution at the end of the eighteenth century; and c) the "Great Acceleration" that began in the 1950s and continues to this day (Fig. 3).46,47
The microbiota historically associated with Homo sapiens has changed over time, as has its vertical transmission from parents to children. This chronological chart shows the successive stages, the main factors and the potential consequences of the changes. 1: Rate of infectious diseases: 2: Rate of allergic and autoimmune diseases; 3: Pasteurisation and new food preservation systems; 4: Use of antibiotics; 5: Physical activity; 6: Use of baby formula; 7: Consumption of processed foods and/or foods high in fat, sugar and/or salt; 8: Caesarean section rate; 9: Diversity of the human microbiota. The stars represent major changes in gut microbiota composition. The bigger the star, the greater the change (original design by Juan Miguel Rodríguez).
The Neolithic period was a gradual process in which humanity moved from a hunter-gatherer economy to planned systems of food production thanks to the domestication of plant, animal and microbial species. A more stable and continuous supply of food reduced the diversity of the Neolithic diet and of the gut microbiota. Analysis of microbiomes preserved in coprolites has confirmed that diet drastically affected the composition of the microbiome, as "Neolithic" microbiomes are similar to those of modern rural communities that maintain fibre- and polysaccharide-rich diets48–50, but different to the microbiomes of the hunter-gatherer tribes that have endured to this day.51
The industrial revolution triggered the second great change in human microbial colonisation, as communities moved from a rural economy to an urban, industrialised and mechanised economy. The great rural exodus to the cities created entire neighbourhoods without any basic health infrastructure that were densely populated by people with limited access to fresh food. Tuberculosis, diphtheria, whooping cough, measles, smallpox, cholera, typhoid fever and many other infectious diseases spread quickly, resulting in high mortality rates.
Pasteur and Koch found that the infections were caused by microorganisms, and Joseph Lister and others pioneered the use of aseptic and antiseptic methods in surgery and medicine. Medical and pharmacological advances (antibiotics), together with public health measures to provide a supply of drinking water, prevent food contamination, etc., successfully controlled the great infectious plagues that devastated our ancestors just a few decades ago. But the new health measures triggered the third great, perhaps irreversible change in modern human microbial colonisation.52 Its consequences can be seen in the growing incidence of modern plagues: chronic non-communicable diseases.52
Hygiene hypothesisThe hypothesis was proposed by Strachan in 1989, having observed an inverse correlation between hay fever and number of older siblings in a population of 17,000 British citizens born in 1958.53 Based on the assumption that more children at home means more shared "germs", Strachan concluded that infections of early childhood protect against allergies. The word "hygiene" only appeared in the title of the article but that was enough to coin the name of the hypothesis. Later, Bach54 proposed that the hypothesis could be extended to also incorporate autoimmune diseases. A scientific explanation was even found to endorse the hygiene hypothesis: reduced or no contact with infectious agents decreases Th1 activity (instrumental in response to bacterial infections), which results in increased Th2 cell activity, a hallmark of allergic disorders.
Old friends hypothesisEpidemiological studies began to break the assumed link between infections and reduced allergy risk: not only do measles and other infections that affect the respiratory mucosa not protect against allergic diseases, but they actually increase the risk in many cases.55 Neglecting personal hygiene would not have any significant impact on the incidence of chronic inflammatory and allergic disorders. Rather, the number of infections would increase. Epidemiological and experimental evidence supports a different hypothesis, known as the "old friends hypothesis".56 According to this hypothesis, to promote the proper development of the immune system, it is not pathogen infections in early childhood that are important, but early and regular exposure to a wide range of harmless microbes or "old friends". These can be found throughout human history and are recognised by the immune system, and train it to react appropriately to various stimuli and threats. The sanitary improvements gradually introduced from the nineteenth century onwards reduced our exposure to the microbes that would have historically lived in symbiosis with humans (Fig. 3).
Globally, changes to the microbiota of people living in industrialised countries are the result of multiple factors. Of these, diet and excessive use of antibiotics deserve special mention as they offer opportunities to improve the course of events. Despite the great success of antibiotics in fighting infections, their intensive use has caused two major problems: a) the spread of multidrug-resistant bacteria, particularly from the 1980s onwards, which is currently considered one of the biggest threats facing public health57, and b) their negative and indiscriminate impact on the microbiome57,58. Many aspects of human physiology, from immune defence or metabolism to behaviour, seem to be altered if a wide range of microbial genes are lacking. Epidemiological and experimental studies have found a positive correlation between exposure to antibiotics and the onset of practically all "modern plagues", which first emerged in the 1940s, coinciding with the widespread roll-out and use of antibiotics.58 It is important to note that antibiotic use per capita is greatest in infants under the age of two years.59
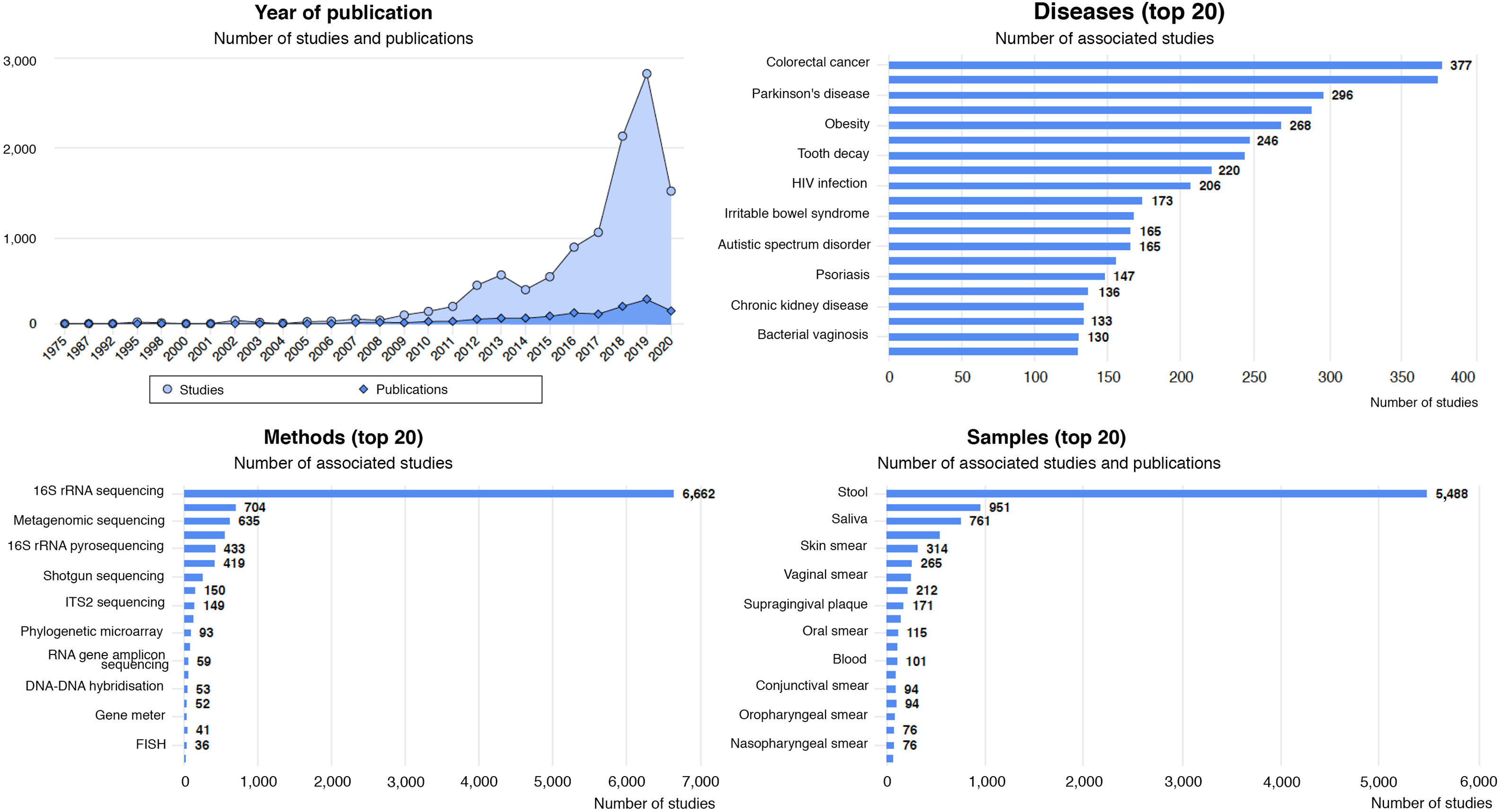
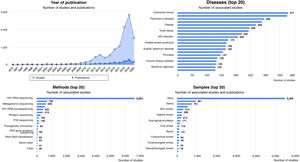
Diseases associated with gut dysbiosisDysbiosis of the gut microbiota has been associated with a long list of inflammatory, autoimmune, metabolic and neoplastic diseases, and with some behavioural disorders. The Disbiome® database (https://disbiome.ugent.be)60 is an archive of studies on the microbiota-disease relationship for more than 300 different diseases (Fig. 4). In most of these, the scientific evidence is not sufficient to ascertain whether dysbiosis precedes the disease or whether the disease itself and its treatment cause dysbiosis.42 Nevertheless, experimental faecal microbiota transplant (FMT) models suggest that there is some form of causal relationship between the altered microbial community and certain diseases.26 This section reviews those diseases with evidence of gut dysbiosis.
Statistical graphs from the Disbiome® website depicting publications and studies that associate changes in microbiota composition with human diseases (https://disbiome.ugent.be; reference Janssens et al.60).
Despite improvements in neonatal intensive care, the number of children with necrotising enterocolitis (NE) is increasing, due to the greater survival rates of very-low-birth-weight newborns. In most cases, the disease manifests without warning signs and continues to be devastating, with high mortality rates and numerous complications. In survivors, post-operative short-bowel syndrome entails significant morbidity: dependence on parenteral nutrition, recurrent infections, poor growth, liver failure, prolonged hospital stays and long-term neurological development disorders.
Prematurity is the main risk factor. Poor gut microbiota implantation plays a key role in its pathogenesis.61 The structure of the microbial community in the faeces of very-low-birth-weight newborns differs significantly between those who may go on to develop NE and those who will not, particularly among babies born before 27 weeks of gestation.62 These differences become clear one month after birth and precede the onset of the disease. An increase in gammaproteobacteria (Enterobacteriaceae) that cause inflammation and a decrease in SCFA-producing anaerobes seem to be the microbial signature that precedes NE.
Its causes are not yet fully understood, and the gammaproteobacteria strains from babies that develop the disease are being studied. An abundance of Bifidobacterium and Lactobacillus seems to be key to protecting the newborn gut. NE may be caused by an inflammatory response secondary to excess Enterobacteriaceae. Modulating the gut microbiota of premature babies through breast milk and dietary supplements reduces the risk of NE. Further studies are required to identify the ideal microbial composition and the most effective combination of supplements.63
Neonatal sepsisThe neonatal gut microbiota undergoes dynamic changes in response to many nutritional and environmental variables. Neonatal infection is a common cause of infant mortality around the world, particularly in very premature babies. Sepsis is defined as life-threatening organ dysfunction caused by a dysregulated host response to microbial infection. Preterm babies often exhibit neonatal dysbiosis, which typically involves the excessive growth of a single species of the facultative anaerobic organisms (Enterococcaceae, Staphylococcaceae or Enterobacteriaceae). The disease is caused by the systemic spread of gut microorganisms.
The prolonged use of broad-spectrum antibiotics in premature newborns has been associated with a higher risk of sepsis. An experimental study64 reviewed mechanisms to prevent the expansion of pathobionts, commensals that can systemically spread, such as Klebsiella pneumoniae. In the murine model, maternal exposure to various antibiotics that eradicated or enriched the population of Lactobacillus murinus exacerbated or prevented sepsis, respectively, and the prophylactic administration of some, but not all, lactobacilli, prevented sepsis. The administration of E. coli was also effective due to nutritional competition with Klebsiella. The gut oxygen level seems to be key to the colonisation dynamic, promoting dysbiosis and sepsis. The pathways that restrict oxygen in the lumen in adults are not operational in newborns.
Protein energy malnutritionMalnutrition affects more than one billion people worldwide and is a significant cause of mortality. In many cases, malnutrition is associated with diarrhoea and intestinal inflammation (colitis), which further increases morbidity and mortality. It has recently been suggested that the gut microbiota may play a modulating role in this process. In fact, malnutrition and tryptophan depletion seem to be influential in the development of diarrhoea and colitis. Dietary tryptophan is primarily absorbed via the B0AT1/ACE2 transport pathway on the luminal surface of small intestinal epithelial cells. This leads to the activation of mTOR (a factor that regulates antimicrobial peptide expression), either directly by nutrient detection or via the tryptophan-nicotinamide pathway, which significantly affects the composition of the microbiota.65 In the event of intestinal injury, an altered microbiome exacerbates the severity of colitis.
Atopic diseasesAsthma and atopy, classically associated with the hyperactivation of T helper 2 (Th2) adaptive immunity, are common chronic diseases. Asthma afflicts more than 235 million people worldwide.66 Recent evidence has correlated atopy and asthma with microbiome composition and function. Gut microbiota diversity is lower in six-month-old babies who go on to develop food sensitivity at one year67 and wheezing at five years68. An abundance of Clostridia and other genera of the phylum Firmicutes during early childhood is associated with a greater likelihood that milk protein allergy will resolve before the age of eight years.67 The development of allergies, particularly respiratory allergies, is associated with a greater faecal abundance of Ruminococcus gnavus, while levels of this bacteria tend to be low in non-allergic individuals. Increased levels of R. gnavus have been observed before the onset of respiratory allergies concomitant with atopic eczema.69 SCFAs, and particularly butyrate, may promote tolerance. The microbiome of babies who develop allergic sensitivity in childhood lacks genes that encode key enzymes for carbohydrate metabolism and butyrate production.70
Prevalent metabolic disease and cognitive impairmentObese subjects and/or subjects with type 2 diabetes mellitus (DM2) develop cognitive impairment.71,72 A meta-analysis found that obesity is linked to a significantly higher risk of cognitive impairment in adults.73 Cognitive impairment rates among patients with DM2 are 1.5 to 2 times higher than in patients without DM2. Both increased adiposity and obesity-related metabolic dysfunction (abnormal glucose tolerance and insulin resistance) could independently affect cognition.74
Obese individuals, people with DM2 and subjects with cognitive impairment share similar pathogenic mechanisms: low-grade systemic inflammation, insulin resistance, increased body iron stores, oxidative stress, mitochondrial dysfunction, vascular changes (higher arterial blood pressure) and sleep apnoea, which can affect cognition.75 There is evidence suggesting that low-grade chronic inflammation directly damages neurons (decreased neurogenesis), irrespective of its effects on metabolism.76 This results in poorer performance in memory and executive function tests and reduced grey matter and white matter volume, all directly correlated to body adiposity.77 Some meta-analyses have even found that adults with DM2 have markedly impaired motor function, executive function, processing speed, verbal memory and visual memory.78 Memory, executive function and the structures of the medial temporal lobe seem to be particularly affected in obese patients.74 Iron accumulation in normal brain ageing is a common finding associated with cognitive impairment79,80, a process that is exacerbated in obese individuals, particularly in the medial temporal lobe and the hippocampus.80,81
The altered composition of the gut microbiota could play an important role in triggering metabolic inflammation, insulin resistance, DM2 and cognitive impairment.82,83 In axenic (germ-free) models, a lack of gut microbes confers protection against the onset of obesity and cognitive learning and memory deficits, supporting the role of the microbiota on higher brain functions.83–85 One study found that recent memory impairment in obese subjects was associated with aromatic amino acid metabolism by the gut microbiota.86
Diabetes mellitusDiet is a key factor in gut microbiota composition, and there is increasing recognition of how different dietary microbial metabolites are associated with insulin secretion, insulin sensitivity and the incidence of type 2 diabetes.87 By way of example, butyrate is produced by specific dietary fibre-fermenting bacteria. The relative loss of these bacteria has been consistently recorded in both prediabetes and type 2 diabetes in different cohorts and different ethnic groups.88 Glycaemic response to a meal is actually determined both by the physiology of the host as well as by the composition of the gut microbiota.89
Inflammatory bowel diseaseUlcerative colitis and Crohn's disease are characterised by exaggerated immune-inflammatory responses to non-pathogenic microorganisms of the gut microbiota, generating ulcerative lesions that become chronic. Several studies have found marked deficiencies in the composition of the faecal and mucosal microbiota.90,91 Patients exhibit a loss of microbial richness and diversity compared to healthy subjects, with fewer butyrate producers (Faecalibacterium, Roseburia, Coprococcus, Lachnospira, etc.) and decreased Akkermansia muciniphila, which is a typical coloniser of the intestinal mucosa, coupled with an overgrowth of proteobacteria, fusobacteria and streptococci. Loss of diversity is particularly prevalent during and after flare-ups.91 Disruption of the symbiotic balance due to poor immune-inflammatory control of the gut microbial population leads to the selection of oxygen-resistant species with inflammatory potential.
A balanced microbial ecosystem is not restored in patients with ulcerative colitis and Crohn's disease during periods of remission, and variability in the microbial composition due to ecosystem instability has been observed. The greater the microbial instability, the greater the risk of a new flare-up.92
Functional digestive disordersIrritable bowel syndrome is the most common functional disorder and typically manifests with recurrent abdominal pain associated with irregular bowel movements, either in the form of constipation or diarrhoea. Patients exhibit quantitative changes in gut bacterial species compared to the healthy population. Patients with diarrhoea have fewer butyrate producers like F. prausnitzii and lower microbial diversity, while methanogen numbers are higher in constipated patients.93
In subjects with functional disorders due to excess gas, food-induced symptoms, such as bloating, distension and pain, were associated with microbial instability. A diet rich in fermentable vegetables induces symptoms in patients, while the composition of the main phyla and genera of the gut microbiota changes abruptly with the onset of symptoms. Healthy subjects show no symptoms and their gut microbiota remains stable.94 These data suggest that the functional competence of the gut microbiota confers stability and prevents the onset of postprandial abdominal symptoms.
Immunosenescence and frailtyThe term "immunosenescence" refers to changes to our immune system as we get older. These changes are in essence the progressive exacerbation of the inflammatory tone and reduced capacity to generate adaptive responses to new antigens. Increased serum inflammatory markers (TNF-〈, IL-6, IL-8, CRP) and worsening frailty and comorbidity indices have been found to be correlated with progressively lower gut microbiota richness and diversity during the ageing process.15 Reduced gut microbial diversity typically involves fewer butyrate-producing species and a relative increase in the number of proteobacteria with inflammatory potential. Insufficient consumption of fruit and vegetables could explain these changes15, given that an interventional study with a Mediterranean diet successfully improved the gut microbiota composition, reversed the inflammatory changes and reduced the frailty indices.95
Graft versus host diseaseHaematopoietic stem cell transplantation is the treatment of choice for several diseases, particularly malignant and non-malignant blood diseases. Its efficacy is limited by serious, life-threatening complications, such as the incidence of untreatable infections during immunosuppression and graft versus host disease. The role of the gut microbiota in terms of "resetting" the immune system through haematopoietic stem cell transplantation and its potential impact on the onset of complications is currently being studied.
Poor faecal microbiota diversity has been associated with significantly higher mortality (52%) versus high microbiota diversity (8%), while abundance of Enterobacteriaceae (Escherichia, Proteus, Klebsiella, etc.) has been correlated with a higher incidence of bacteraemia and lower overall survival.96 Poor bacterial diversity in faeces is also a predictor of a fatal outcome in graft versus host disease.96 Antibiotics considerably reduce gut microbiota diversity, and transplant recipients typically receive multiple antibiotic treatments. The use of antibiotics against anaerobes or colonisation by multidrug-resistant bacteria, which requires frequent or prolonged antibiotic exposure, has been found to be associated with a higher incidence of graft versus host disease.96
These observations suggest that it may be beneficial to alter the gut microbiota of transplant recipients in order to prevent or mitigate complications and improve survival rates. An uncontrolled series of patients with multidrug-resistant bacteria found that FMT before or after haematopoietic stem cell transplantation is an effective and safe multidrug-resistant bacterial decolonisation strategy.97 Although further studies are clearly needed to confirm the efficacy of FMT in preventing complications in transplant recipients, it is nevertheless a promising therapeutic strategy.
Functional modulation and interventionModulating the gut microbiota to improve human health is the ongoing subject of intensive and extensive research that is testing strategies such as dietary intervention with various nutrients, including prebiotics, probiotics, synbiotics and other related strategies (postbiotics, paraprobiotics), or FMT. Most likely, an understanding of the individual composition of the gut microbiota will facilitate future personalised nutrition protocols for optimal health. For the time being, the main objectives are to increase microbial diversity and enhance the functional capacity to generate SCFAs, particularly butyrate.
DietNutrients are not just vital to human health, but also to the health of the gut microbiota. The metabolic functions performed by the gut microbiota are linked to the digestion of complex polysaccharides, SCFA production, bile acid metabolism, vitamin production, etc., with the resulting trophic effects on the intestinal barrier and immune system. As such, diet is key to achieving and maintaining symbiosis between the microbiota and the host, as it determines and modulates the establishment of the gut microbiota in childhood, and its structure and functions in adulthood. Other relevant factors in addition to diet (food components, nutrients, eating patterns, etc.) include medicines, hygiene, circadian rhythm, intermittent fasting, seasonal changes and industrialisation, among others.98
Enterotypes seem to be influenced by the host's routine eating pattern.99 The most common enterotype in industrialised countries is Bacteroides and is associated with the eating habits of the urban population. The most common enterotype in agricultural regions is Prevotella, with high-fibre diets low in protein and animal fats.48,100 Other studies101,102 have identified seasonal changes in the microbiota, consistent with increased consumption of fresh fruit and vegetables in the summer, compared to frozen or tinned foods in the winter. Dietary differences between populations could explain the taxonomic variability of the gut microbial ecosystem.
Short-term dietary interventions could induce rapid changes in microbiota composition associated with drastic changes in dietary fibre intake. The extent of the effects is relatively modest compared to interindividual variability in microbial taxonomy, and changes are not consistent from one individual to another, meaning that everyone changes in a different way.103 Increased protein and animal fat together with a lack of dietary fibre has been found to increase the abundance of bile salt-tolerant microorganisms (Alistipes, Bilophila) and to reduce levels of Firmicutes that metabolise vegetable complex polysaccharides (Roseburia, Eubacterium and Ruminococcus). In contrast, the abundant consumption of dietary fibre, fruit, vegetables and other plant-based foods is associated with important and significant increases in fermentative species. Controlled studies have shown that the consumption of resistant starch or other indigestible polysaccharides increases the abundance of fermentative species like Ruminococcus bromii and Eubacterium rectale.104
In experimental models, high fat intake can imbalance the gut ecosystem and contribute to the onset of inflammatory processes. The mechanism seems to be mediated by increased lipopolysaccharide absorption, subclinical endotoxaemia and TLR4 activation. This same mechanism is believed to contribute to the development of cardiovascular and metabolic diseases.105 The microbiota may also help to regulate lipid and glucose metabolism thanks to the interaction of bile acids in the terminal ileum and modulation of the G protein-coupled bile acid receptor 1 (TGR5). This evidence raises questions about the role the microbiota plays in the individual efficacy and safety of some of the treatments tested with bile acids in obesity.106
Proteins also modulate microbiota composition and metabolite production. Microbial catabolism of amino acids generates indoles, phenols, ammonia and amines, which can combine with nitric oxide to form genotoxic nitroso compounds that are associated with colorectal carcinogenesis.107 In contrast, indolepropionic acid and indole-3-acetate deriving from the microbial metabolism of tryptophan maintain intestinal homeostasis, protect from experimental colitis and/or reduce hepatocyte inflammation.106 The role of microbes in the generation of TMAO (a cardiovascular risk factor23,24) from carnitine and phosphatidylcholine has already been mentioned above.
Although the impact of food additives on the microbiota and intestinal homeostasis has not been well studied, it is known that some food emulsifiers, such as polysorbate-80 or carboxymethyl cellulose, trigger metabolic abnormalities in obese mice. Similarly, non-caloric sweeteners can induce glucose intolerance by interacting with the microbiota, both in animal models and in humans.106
Vegetarian or vegan diets are rich in complex carbohydrates and would therefore be expected to have a beneficial impact on the gut microbiota. Some cross-sectional and interventional studies have identified changes in different taxa but only minimal changes in terms of richness and diversity. These minimal changes may be enough to justify the benefits in SCFA production, which is higher in the vegetarian population. The benefits of vegetarian diets are also likely to derive from the intake of phytochemicals, such as isoflavones, which were mentioned above, and others.106
The Mediterranean diet has demonstrated enormous benefits in population health, reducing the risk of mortality and many chronic diseases.108,109 Studies suggest that dietary intervention should prioritise the inclusion of a wide variety of plant-based foods in significant quantities, rather than excluding animal-based foods, which supports the concept that a varied and diverse diet promotes microbiota stability.
PrebioticsAlthough a bifidus factor in breast milk was identified as being responsible for the increase in bifidobacteria in children as early as the mid-twentieth century, the term "prebiotic" only appeared in print for the first time in 1995.110 Gibson and Roberfroid defined the concept as "a non-digestible food ingredient that beneficially affects the host by selectively stimulating the growth and/or activity of one or a limited number of bacteria in the colon, and thus improves host health." According to this definition, only certain carbohydrates, such as galacto-oligosaccharides (GOS), lactulose, inulin and fructo-oligosaccharides (FOS) meet the criteria. Other compounds that may meet these criteria are isomalto-oligosaccharides (IMO), xylo-oligosaccharides (XOS), transgalacto-oligosaccharides (TOS) and soybean oligosaccharides (SBOS). They are found in milk, vegetables and leafy vegetables, fruit, grains, pulses and nuts.99
In 2017, the International Scientific Association for Probiotics and Prebiotics (ISAPP) revised the concept and proposed that a prebiotic is "a substrate that is selectively utilized by host microorganisms conferring a health benefit".111 The new concept would apply to different substances, including carbohydrates, polyphenols, polyunsaturated fatty acids, etc.
The consensus document retained the requirement for selective microbiota-mediated mechanisms, and that beneficial health effects must be demonstrated by well-designed clinical trials. Scientific and clinical advances in the field of molecular techniques are showing that the selective use of prebiotics is not exclusive to lactobacilli and bifidobacteria but extends to genera such as Eubacterium and Roseburia. Although most prebiotics are administered orally, the panel acknowledges that they can also be administered to other parts of the body colonised by microorganisms, such as the skin or vaginal tract.
Special mention should be made of human milk oligosaccharides (HMOs), an extraordinarily complex mixture with more than 1000 chemical structures described, which act as more than mere substrates for the gut microbiota and are involved in numerous functions that are beneficial to local and systemic infant health.5
Their potential for use in obesity, insulin resistance, hepatic steatosis, anxiety and depression is currently being studied. The evidence generated in different diseases supports the hypothesis that prebiotics play a health-promoting role in a healthy population, which can be detected through extensive observational studies.111 Resistant starch and non-carbohydrate prebiotics are emerging prebiotics currently under study.
ProbioticsMicroorganism-fermented foods such as bread, beer, wine, kefir, yoghurt, kumis and cheese have been consumed by humans since the Neolithic period, for both their nutritional and therapeutic benefits. A commission of experts convened in 2001 jointly by the Food and Agriculture Organization (FAO) and the World Health Organization (WHO) defined probiotics as "live microorganisms that, when administered in adequate amounts, confer a benefit to the health of the host".112 This definition has been widely accepted by the international scientific community and has been acknowledged in the consensus documents of the Sociedad Española de Microbiota, Probióticos y Prebióticos (SEMIPyP) [Spanish Society of Microbiota, Probiotics and Prebiotics]113 and the ISAPP.114 The term "next-generation probiotics" is sometimes used to refer to those beneficial species that make up part of the human gut microbiota (Faecalibacterium prausnitzii or Roseburia intestinalis). Other terms have also been proposed to denote probiotics with indications in specific areas, such as "psychobiotics" and "oncobiotics". The definition of probiotics does not include dead microorganisms or components or substances produced by microorganisms, even if they demonstrate healthy biological effects. These are known as "paraprobiotics" and "postbiotics", respectively.
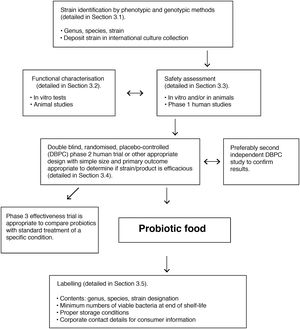
The SEMIPyP and ISAPP consensus documents stressed that probiotics should be perfectly characterised in terms of their strain, and their viability must be retained for the entire shelf life of the products in which they are supplied. Strict quality systems should be implemented that guarantee their taxonomy and the presence of sufficient quantities of viable microorganisms.
Microorganisms marketed as probiotics include yeasts (Saccharomyces, Kluyveromyces) and bacteria of various genera (Lactobacillus, Streptococcus, Enterococcus, Pediococcus, Bifidobacterium, Propionibacterium, Bacillus, Escherichia). Precise taxonomic classification of the microorganism is vital in order to assess its safety. Moreover, the probiotic concept only applies to strains that have been evaluated in accordance with the FAO/WHO guidelines (Fig. 5).112 All probiotics must be identified by their genus, species, subspecies (if applicable) and an alphanumeric strain designation. The health benefit must be demonstrated through randomised, controlled clinical trials. In addition, the proven benefit for a specific health condition (e.g. acute diarrhoea) cannot be extrapolated to other indications (e.g. allergy).
FAO/WHO guidelines for evaluation of a microorganism as a probiotic (reference Roberfroid et al.110).
Several clinical trials have evaluated the efficacy and safety of various probiotics for different indications, including the prevention and treatment of acute diarrhoea, antibiotic-associated diarrhoea, mild or moderate functional digestive symptoms, lactose intolerance, infant colic, etc. The World Gastroenterology Organisation (WGO) published an open-access practice guideline containing information about strains, posology and level of evidence for various gastrointestinal applications.115 The potential therapeutic use of probiotics in other diseases (e.g. obesity, insulin resistance, hepatic steatosis, anxiety or depression) is currently being studied and should be viewed with caution. Probiotics may help to promote health in a healthy population.114,116
SynbioticsA synbiotic is a product that combines at least one probiotic and one prebiotic. A product can only be called synbiotic if it is perfectly characterised and has been proven to induce a beneficial effect greater than the sum of those generated separately by its component parts.117 A popular example of this type of product is the combination of microorganisms of the genus Bifidobacterium or Lactobacillus with fructo-oligosaccharides.116
Faecal microbiota transplantFaecal microbiota transplant (FMT) is a method that aims to change a patient's gut microbiota in order to normalise its composition and obtain a therapeutic benefit. It involves infusing a stool suspension donated by a healthy individual into the digestive tract of the recipient. Although this technique was used in China as early as the fourth century, it gained very little traction throughout history until its extraordinary efficacy in treating refractory and recurrent Clostridioides difficile-associated diarrhoea was demonstrated in 2013, with resolution rates in excess of 90%.118
The donor must be carefully selected to avoid adverse effects, such as the inadvertent transplantation of multidrug-resistant bacteria. Experience accrued to date suggests that the procedure entails no significant side effects when performed in accordance with agreed protocols, such as the European protocol119 and the protocol issued by the Societat Catalana de Digestologia [Catalan Society of Digestology] and the Societat Catalana de Malalties Infeccioses i Microbiologia Clínica [Catalan Society of Infectious Diseases and Clinical Microbiology], which is more up-to-date120. Its application in other diseases, such as inflammatory bowel disease, metabolic syndrome and haematopoietic stem cell transplantation has also been investigated in recent years.121 Apart from the aforementioned indication to resolve recurrent Clostridioides difficile-associated diarrhoea, this technique is currently only used in research protocols approved by an ethics committee. It is vital to optimise procedures (particularly given the current context of the coronavirus pandemic), better define deficiencies and select active microorganisms that will allow the application of personalised approaches to treat specific diseases.
FundingThe drafting of this article was funded by the Danone Institute (Barcelona).
Conflicts of interestJA, FG and MSP are members of the Danone Institute scientific committee.
Please cite this article as: Álvarez J, Fernández Real JM, Guarner F, Guarner F, Gueimonde M, Rodríguez JM, et al., Microbiota intestinal y salud, Gastroenterol Hepatol. 2021;44:519–535.