The association between gut microbiota and animal models of multiple sclerosis has been well established; however, studies in humans are scarce.
MethodsWe performed a descriptive, cross-sectional study comparing the relative composition of gut microbiota in 30 patients with multiple sclerosis (15 treated with interferon β-1b, 15 not receiving this treatment) and 14 healthy controls using next generation sequencing.
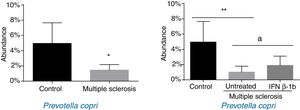
ResultsPatients with multiple sclerosis and controls showed differences in the proportion of Euryarchaeota, Firmicutes, Proteobacteria, Actinobacteria, and Lentisphaerae phyla and in 17 bacterial species. More specifically, we found significant differences in the proportion of Firmicutes, Actinobacteria, and Lentisphaerae and 6 bacteria species between controls and untreated patients; however, these differences disappeared when compared with treated patients. Untreated patients showed a significant reduction in the proportion of Prevotella copri compared to controls, while the bacteria was significantly more abundant in patients treated with interferon β-1b than in untreated patients, with levels resembling those observed in the healthy control group.
ConclusionWe observed differences in gut microbiota composition between patients with multiple sclerosis and controls, and between patients treated and not treated with interferon β-1b. In most cases, no differences were observed between treated patients and healthy controls, particularly for P. copri levels. This suggests that the clinical improvements observed in patients with multiple sclerosis receiving interferon β-1b may result from the effect of the drug on gut microbiota. Longitudinal and functional studies are necessary to establish a causal relationship.
El papel de la microbiota en los modelos animales de esclerosis múltiple está bien establecido; por el contrario, los estudios en humanos son escasos.
MétodosEstudio transversal descriptivo que compara la composición relativa de la microbiota intestinal en 30 pacientes con esclerosis múltiple (15 tratados con interferón β-1b, 15 sin tratamiento) y 14 controles sanos utilizando la secuenciación de última generación.
ResultadosLos sujetos control y los pacientes con esclerosis múltiple presentaron diferente abundancia de los filos Euryarchaeota, Firmicutes, Proteobacteria, Actinobacteria, y Lentisphaerae y 17 especies bacterianas. Concretamente, la abundancia en Firmicutes, Actinobacteria y Lentisphaerae y 6 especies mostró diferencias al comparar los grupos control y sin tratamiento, desapareciendo esta diferencia cuando se compararon con los pacientes tratados. Se observó reducción estadísticamente significativa en la abundancia de Prevotella copri en pacientes sin tratamiento en comparación con controles, mientras que los tratados con interferón β-1b presentaron un aumento significativo frente a pacientes sin tratamiento, asemejándose al grupo de pacientes sanos control.
ConclusiónLa composición de la microbiota intestinal fue diferente entre los pacientes con esclerosis múltiple y los controles, y entre los pacientes sin tratamiento y los tratados con interferón β-1b. En la mayoría de los casos, no se encontraron diferencias entre los pacientes tratados y los controles sanos, siendo especialmente evidente con P. copri. Esto podría indicar que la influencia del interferón β-1b sobre la microbiota intestinal podría subyacer en los beneficios clínicos observados en pacientes con esclerosis múltiple que siguen este tratamiento. Serán necesarios estudios longitudinales y funcionales para poder mostrar causalidad.
Multiple sclerosis (MS) is a demyelinating disease affecting over 2.5 million people globally; like other autoimmune diseases, it has shown rising incidence and prevalence in recent years.1 The pathogenesis of the disease is not fully understood, although it is widely accepted that both genetic predisposition and exposure to certain environmental factors are involved. Established environmental factors include Epstein-Barr virus, smoking, geographical latitude, and vitamin D deficiency; and new environmental factors currently under study include excessive sodium, vitamin A, or alcohol intake, overweight in adolescence, high leptin levels, and lack of exposure to helminths or Helicobacter pylori.1 These new environmental factors may also include the gut microbiota.2–9 The microbiota's role in human physiology is well described. It is involved in immune homeostasis, affecting both the innate and the adaptive immune responses; in the maturation of lymphoid organs; in the production of IgA-producing plasma cells, intestinal γδ T cells, and CD4 T cells; in the genetic expression of toll-like receptors and the major histocompatibility complex type II; and in the production of cytokines; etc. Underlying all these functions is the microbiota's influence over regulatory T cells and T helper cells, and more specifically over the Th1-Th17/Th2 balance, which also explains its influence over susceptibility to infection, inflammatory disease, cancer, and the subject of interest here, autoimmune disease.1
Dysbiosis has been linked to a proinflammatory state associated with numerous autoimmune conditions, including inflammatory bowel disease, type 1 diabetes, and rheumatoid arthritis; gastrointestinal, neurological, respiratory, metabolic, hepatic, and cardiovascular diseases10; HIV11; and MS. More specifically, and relevantly in the case of MS, there is robust scientific evidence of the microbiota's role in the pathogenesis, prevention, and even the treatment of experimental autoimmune encephalomyelitis, an animal model of MS.1 However, few studies have addressed its role in MS in humans, and the existing studies are in early stages of research.2–9 None of these studies has been performed in a Mediterranean or specifically in a Spanish population; and no study has specifically aimed to assess the influence of the microbiota on the treatment of the disease.
Therefore, this study aims to describe the relative composition of bacterial and archaean taxa in the gut microbiota of a sample of patients from Spain and to detect differences between those not being treated and those receiving interferon (IFN) β-1b.
Material and methodsRecruitment of patientsThis cross-sectional study was conducted at Hospital San Pedro de Logroño (Spain) between September 2014 and April 2015. We sought to recruit patients older than 18 years diagnosed with MS (2010 McDonald criteria) who were attended on either an inpatient or an outpatient basis.12 Patients were divided into 2 groups: the untreated group, who had never received any disease-modifying drug; and the treatment group, comprising patients who had been receiving IFN β-1b every 48 hours for at least 9 months. For each group, we selected the first 15 patients meeting all inclusion criteria and no exclusion criterion. Given the lack of consensus on the selection of control subjects,4 we selected 14 healthy volunteers with no personal history of autoimmune disease or family history of MS.
The exclusion criteria were as follows: treatment in the last month with antibiotics, corticosteroids, immunosuppressants, or probiotic-based medicines or nutritional supplements; previous gastrointestinal surgery (except for appendectomy or cholecystectomy); and personal history of inflammatory bowel disease, coeliac disease, chronic pancreatitis, or any malabsorption syndrome.
The study was conducted in accordance with the ethical principles of the Belmont report, the Declaration of Helsinki, and the UNESCO Universal Declaration on the human genome, and was approved by the clinical research ethics committee of La Rioja (ref. CEICLAR PI 176). All participants gave informed consent to be included in the study.
Collection of samples, DNA extraction, 16s ribosomal rDNA sequencing, and bioinformatic analysisFrom each individual we collected a sample of the first stool of the morning, and used the DNeasy Blood and Tissue Kit (QIAGEN, Germany) to extract DNA from the fresh sample (25mg), following the manufacturer's instructions. A Nanodrop 1000 spectrophotometer (ND-1000; Thermo Scientific, USA) was used to determine purity and concentration. For ribosomal DNA (rDNA) sequencing, we amplified the V4 hypervariable region of the 16s rDNA gene using 515-806-R primers with the Illumina MiSeq system (2×250bp), obtaining approximately 100000 reads per sample.13 Bioinformatic analysis was performed by the company Era7 Bioinformatics (Granada, Spain). In this analysis, the FLASH program was used to assemble both reads obtained with the Illumina system. Reads were assigned to taxa according to their similarity to the 16s rRNA sequences included in the Ribosomal Database Project,14 based on the specificity of their taxonomic assignation in the National Center for Biotechnology Information database (https://github.com/ohnosequences/db.rna16s). This process was carried out with a massive BLAST (basic local alignment 116 search tool) with MG7 analysis.15 The subsequent taxonomic assignation involved 2 methods: BBH (best blast hit) at the phylum level, and LCA (lowest common ancestor) at lower taxonomic levels. LCA was based on advanced metagenomic analysis tools similar to the latest version of the MEtaGenome ANalyzer (MEGAN) program.16 We initially studied taxon richness (α-diversity), and subsequently analysed the differences in the taxa identified in each of the study groups (β-diversity).17
Statistical analysisResults are expressed as mean (standard error of the mean [SEM]). Statistical significance was set at P<.05. Categorical variables were analysed with the chi-square test and the Fisher exact test. Normal distribution was assessed using the Shapiro-Wilk test. Intergroup comparisons were performed with the t test when data were normally distributed in both groups, and the Mann-Whitney U test when one group did not display normal distribution. Comparisons between all 3 groups (controls, untreated group, and treatment group) were performed using analysis of variance and with a post hoc multiple-comparisons analysis using the Tukey test, for normally distributed data. Where data were not normally distributed, we compared the 3 groups with the Kruskal-Wallis test with a post hoc multiple-comparisons analysis using the Dunn multiple-comparisons test. Statistical analysis was performed using the SPSS software, version 19.0 (SPSS® Inc., USA) and GraphPad Prism 6 (GraphPad Prism®, USA).
α-Diversity was calculated with 4 indices (Shannon index, Margalef diversity index, Chao1 index, and the alpha index) using R (Vegan: Community Ecology Package, version 2.3.2: http://CRAN.R-project.org/package=vegan). β-Diversity was assessed by performing a comparative analysis of all phyla, the 40 most abundant taxa, and taxa showing differences after a metagenomic analysis performed with the METAGENassist18 web server. These analyses were only performed when reads were obtained from at least 25% of members of each group.
ResultsAll participants were white, and all patients presented relapsing-remitting MS. The epidemiological characteristics of each group are shown in Table 1. The only significant difference identified between groups was in the number of relapses in the past year, which was greater in the group of untreated patients, as we would expect.
Epidemiological characteristics of the sample.
| Controls (n=14) | Untreated (n=15) | IFN β-1b (n=15) | P | |
|---|---|---|---|---|
| Age (years) | 45.2 (3.0) | 39.3 (3.0) | 40.6 (2.3) | .30 |
| Women/men | 7/7 | 12/3 | 9/6 | .23 |
| Smokers/non-smokers | 1/13 | 5/10 | 6/9 | .11 |
| BMI (kg/m2) | 25.1 (0.8) | 24.1 (0.8) | 26.3 (1.2) | .27 |
| MS progression time (months) | 54.7 (14.1) | 59.0 (13.9) | .83 | |
| EDSS score | 1.07 (0.21) | 1.73 (0.38) | .13 | |
| Relapses in the previous year | 0.6 (0.2) | 0.2 (0.1) | .05 | |
| IFN β-1b treatment duration (months) | 42.5 (39.6) |
EDSS: Expanded Disability Status Scale; MS: multiple sclerosis; IFN: interferon; BMI: body mass index; SEM: standard error of the mean.
Data are expressed as mean (SEM).
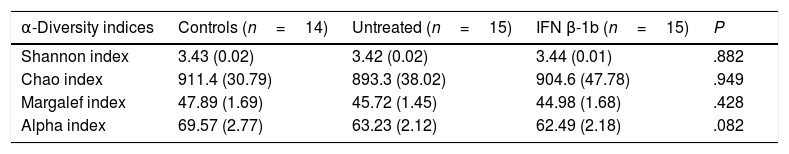
No significant differences were observed between groups with any of the indices used (Table 2), although the alpha index showed a trend towards reduced species richness in patients with MS vs controls (P=.08).
Measures of α-diversity in controls and in patients with multiple sclerosis with and without IFN β-1b treatment.
| α-Diversity indices | Controls (n=14) | Untreated (n=15) | IFN β-1b (n=15) | P |
|---|---|---|---|---|
| Shannon index | 3.43 (0.02) | 3.42 (0.02) | 3.44 (0.01) | .882 |
| Chao index | 911.4 (30.79) | 893.3 (38.02) | 904.6 (47.78) | .949 |
| Margalef index | 47.89 (1.69) | 45.72 (1.45) | 44.98 (1.68) | .428 |
| Alpha index | 69.57 (2.77) | 63.23 (2.12) | 62.49 (2.18) | .082 |
Data are expressed as mean (SEM).
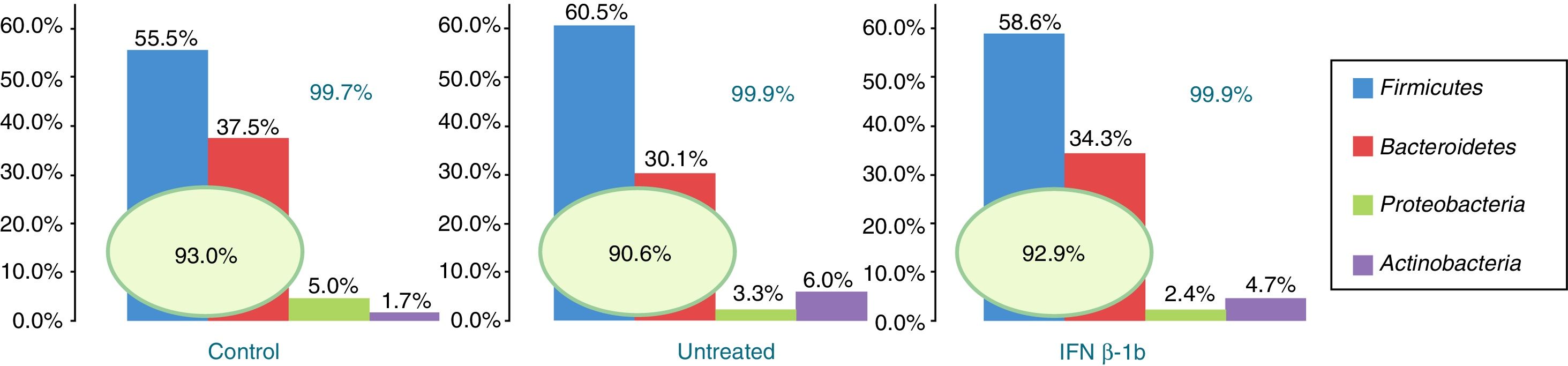
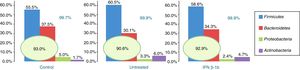
Of the more than 30 bacterial phyla present in the gut microbiota,2 we obtained reads of 12 phyla of bacteria and one archaea phylum. The most abundant phyla in all 3 groups were Firmicutes, Bacteroidetes, Proteobacteria, and Actinobacteria, which accounted for at least 99.70% of the assigned reads. Of these 4 phyla, Firmicutes and Bacteroidetes were the most prevalent, representing over 90% of reads in all groups, followed by Proteobacteria and then Actinobacteria in controls, while the latter phylum was more abundant than Proteobacteria in patients with MS (Fig. 1).
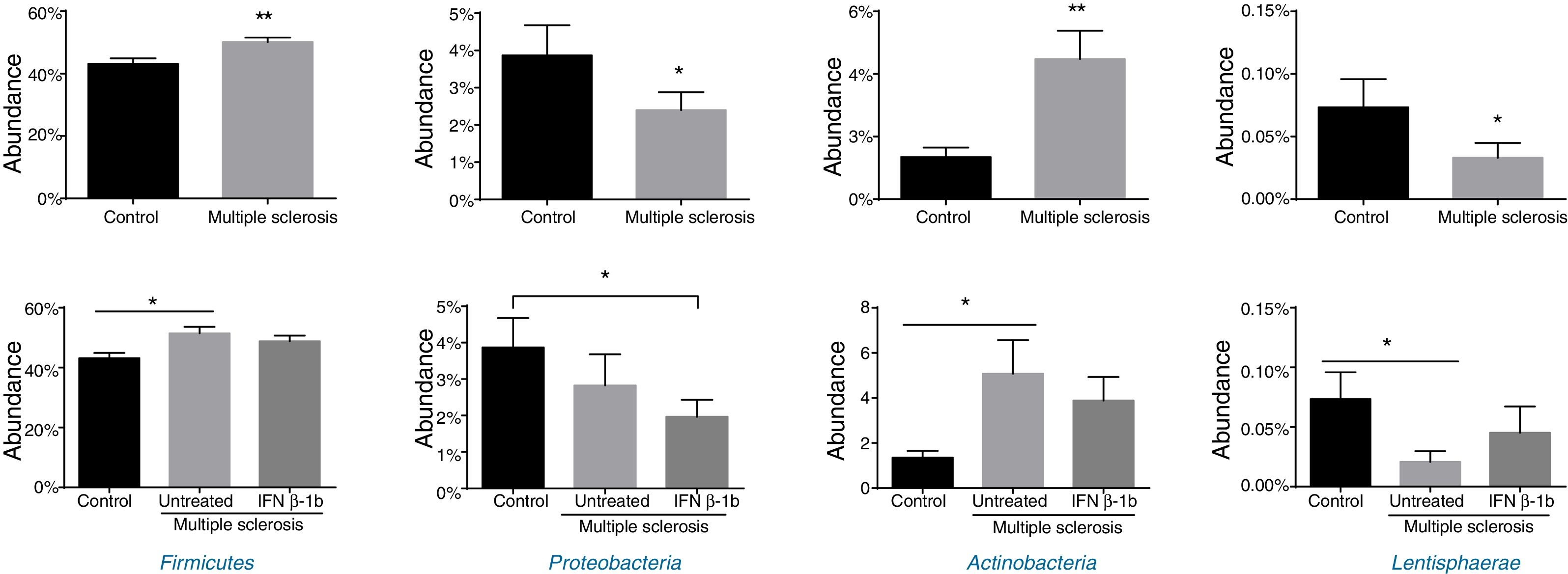
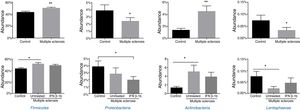
Significant differences were observed between patients and controls in the abundance of the phyla Firmicutes, Proteobacteria, Actinobacteria, and Lentisphaerae (Fig. 2), with patients showing higher levels of Firmicutes and Actinobacteria and lower levels of Proteobacteria and Lentispaerae. When the treatment variable was included in this analysis, differences were detected in the levels of Firmicutes, Actinobacteria, and Lentisphaerae between controls and untreated patients but not between controls and patients in the treatment group. To the contrary, the reduced abundance of Proteobacteria among patients with MS was due to a reduction among treated patients (Fig. 2).
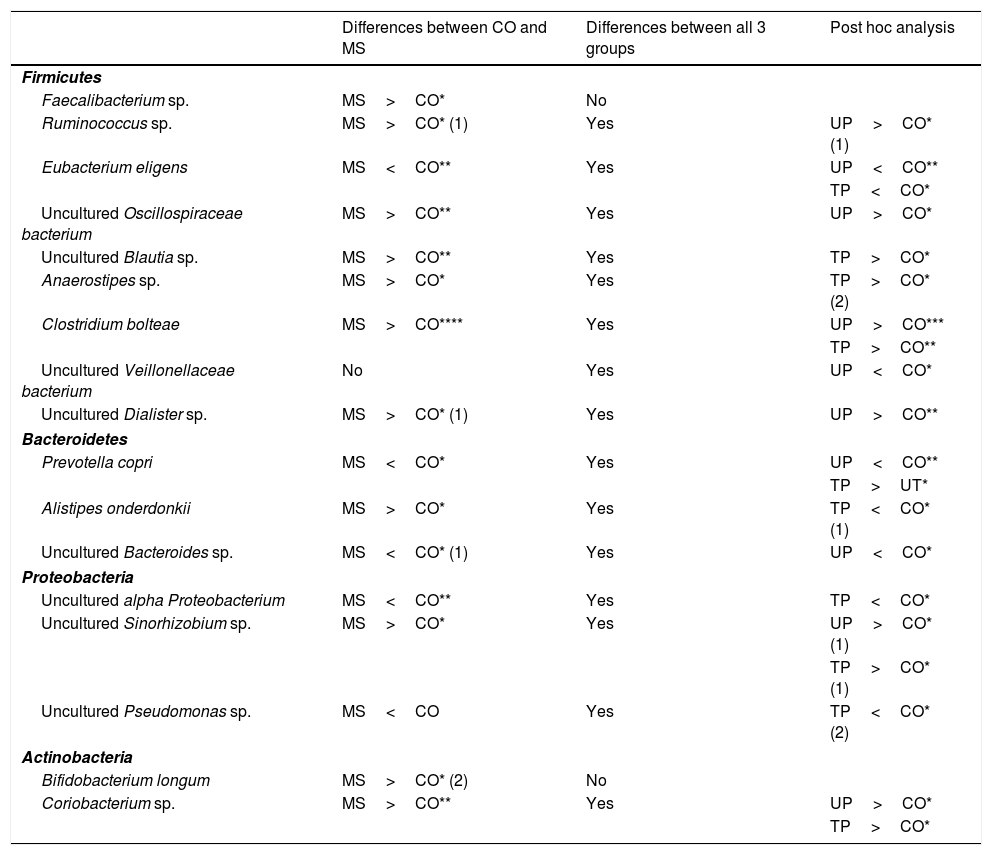
In the analysis of operational taxonomic units (OTU) corresponding to genus and species, 11 OTUs (Faecalibacterium sp.; Ruminococcus sp.; uncultured Oscillospiraceae bacterium; uncultured Blautia sp.; uncultured Sinorhizobium sp.; Anaerostipes sp.; Clostridium bolteae; uncultured Dialister sp.; Alistipes onderdonkii; Bifidobacterium longum; and Coriobacterium sp.) were more abundant in patients with MS than in the control group (Table 3). Seven of these OTUs belonged to the phylum Firmicutes, one to Bacteroidetes, one to Proteobacteria, and 2 to Actinobacteria. On the other hand, 5 OTUs were less abundant among patients with MS (Eubacterium eligens; Prevotella copri; uncultured Bacteroides sp.; uncultured alpha Proteobacterium; and uncultured Pseudomonas sp.). Of these, one belongs to Firmicutes, 2 to Bacteroidetes, and 2 to Proteobacteria (Table 3).
Summary of differences detected in the levels of bacterial species.
| Differences between CO and MS | Differences between all 3 groups | Post hoc analysis | |
|---|---|---|---|
| Firmicutes | |||
| Faecalibacterium sp. | MS>CO* | No | |
| Ruminococcus sp. | MS>CO* (1) | Yes | UP>CO* (1) |
| Eubacterium eligens | MS<CO** | Yes | UP<CO** |
| TP<CO* | |||
| Uncultured Oscillospiraceae bacterium | MS>CO** | Yes | UP>CO* |
| Uncultured Blautia sp. | MS>CO** | Yes | TP>CO* |
| Anaerostipes sp. | MS>CO* | Yes | TP>CO* (2) |
| Clostridium bolteae | MS>CO**** | Yes | UP>CO*** |
| TP>CO** | |||
| Uncultured Veillonellaceae bacterium | No | Yes | UP<CO* |
| Uncultured Dialister sp. | MS>CO* (1) | Yes | UP>CO** |
| Bacteroidetes | |||
| Prevotella copri | MS<CO* | Yes | UP<CO** |
| TP>UT* | |||
| Alistipes onderdonkii | MS>CO* | Yes | TP<CO* (1) |
| Uncultured Bacteroides sp. | MS<CO* (1) | Yes | UP<CO* |
| Proteobacteria | |||
| Uncultured alpha Proteobacterium | MS<CO** | Yes | TP<CO* |
| Uncultured Sinorhizobium sp. | MS>CO* | Yes | UP>CO* (1) |
| TP>CO* (1) | |||
| Uncultured Pseudomonas sp. | MS<CO | Yes | TP<CO* (2) |
| Actinobacteria | |||
| Bifidobacterium longum | MS>CO* (2) | No | |
| Coriobacterium sp. | MS>CO** | Yes | UP>CO* |
| TP>CO* | |||
Observations: (1) Difference was only statistically significant when atypical values were removed; (2) difference was no longer significant when atypical values were removed. All remaining differences were statistically significant both with and without atypical values.
CO: controls; MS: patients with MS; TP: treated patients; UP: untreated patients.
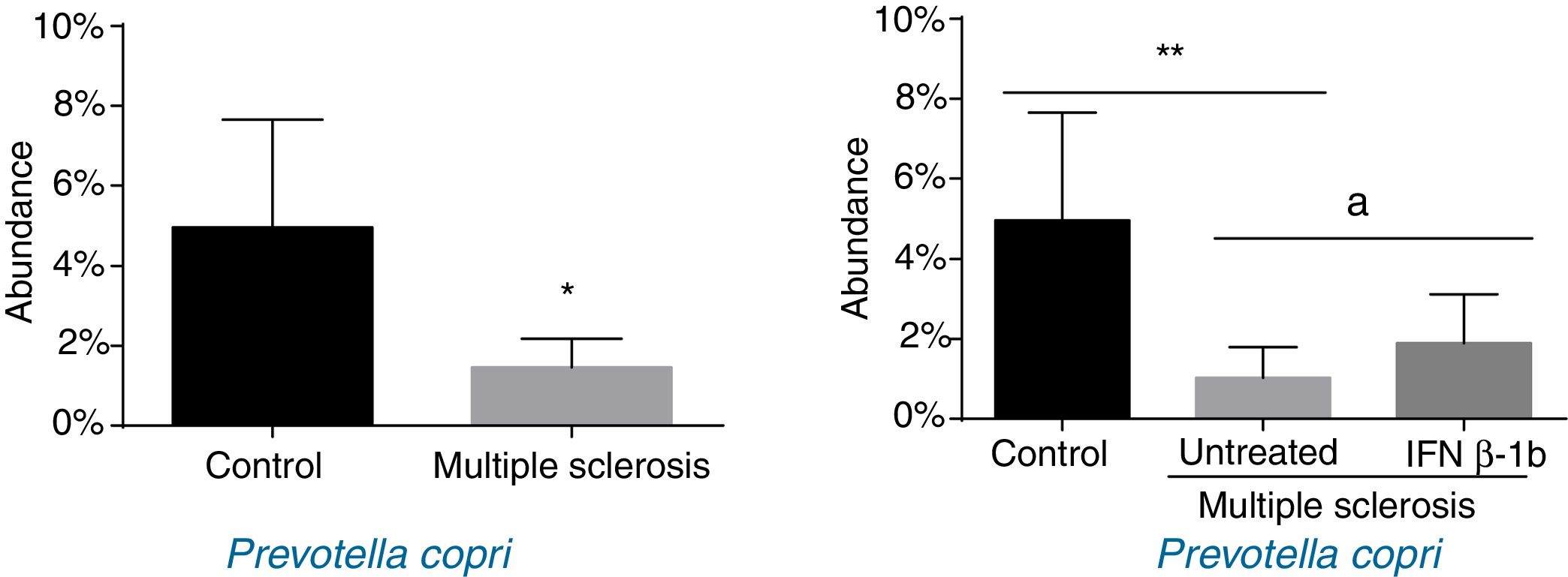
When patients were compared by treatment group, we only observed significant differences for P. copri. Significantly lower levels were observed in untreated patients than in controls (P<.01), and the IFN β-1b group showed greater abundance than untreated patients (P<.05) and similar levels to controls (Fig. 3). The difference between patients and controls was also observed at the genus level, with abundance of Prevotella being far lower among patients than in controls (0.041% [0.038%] vs 0.003% [0.001%]; P<.05).
Seven of the 9 Firmicutes OTUs that were found in greater abundance in patients with MS than among controls belonged to the order Clostridiales (Supplementary Material Fig. S1). With the exception of Faecalibacterium sp., all Clostridiales OTUs showed significant differences between the 3 groups. The other 2 Firmicutes species in which we found significant differences (uncultured Veillonellaceae bacterium and uncultured Dialister sp.) belong to the order Selenomonadales (Table 3). Both species showed interesting results for the comparison of all 3 study groups, with significant differences in abundance between untreated patients and controls accounting for the differences observed in the comparison between the group of all patients and controls. Patients receiving IFN β-1b showed similar abundance to controls; differences were also observed in comparison to untreated patients, although they were not statistically significant (Supplementary Material Fig. S2).
As well as in P. copri, we also observed significant differences in 2 OTUs from the Bacteroidetes phylum (A. onderdonkii and uncultured Bacteroides sp.; Supplementary Material Fig. S3), 3 from Proteobacteria (uncultured alpha Proteobacterium, uncultured Pseudomonas sp., and uncultured Sinorhizobium sp.; Supplementary Material Fig. S4), and 2 from Actinobacteria (B. longum and Coriobacterium sp.; Supplementary Material Fig. S5).
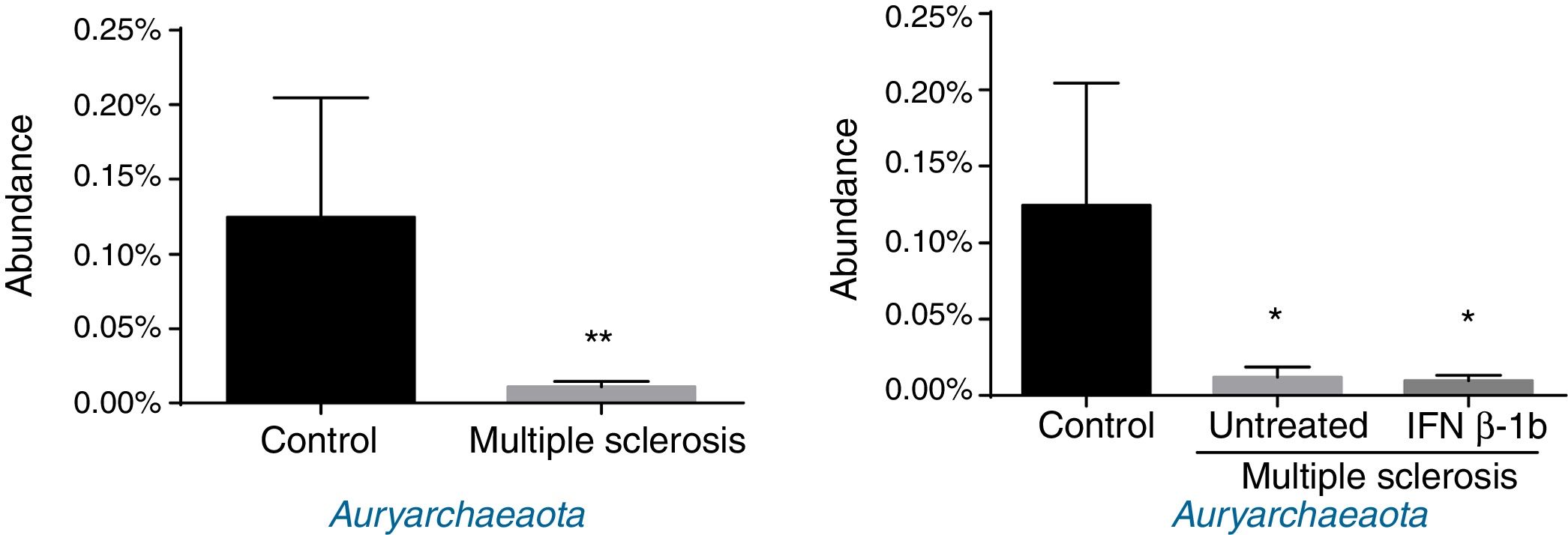
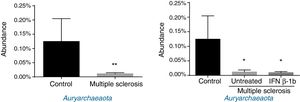
Regarding archaea, significant differences were observed in abundance of the phylum Euryarchaeota (all reads belonged to this phylum and corresponded to a single genus, Methanobrevibacter). Lower abundance was observed in patients than among controls, independently of treatment with IFN β-1b (Fig. 4).
DiscussionThis study is one of the few published works reporting differences in microbiota composition between patients with MS and controls, both at the phylum level and at lower taxonomic levels.2,18 Our findings are of great interest, as the modulation and restoration of the microbiota has been proposed as a promising strategy for the treatment of MS. In this context, our results demonstrate that patients receiving the disease-modifying drug IFN β-1b and healthy controls show similar abundance of various taxa that are altered in patients with untreated MS. These data raise the question of whether the clinical improvement observed in patients receiving the drug may to some extent be mediated by its effect on the microbiota.
The bacterial richness (α-diversity) of the microbiota is altered in numerous clinical situations, including obesity and HIV.11,19 However, we observed similar bacterial richness in the microbiota of patients with MS and in healthy controls, independently of treatment with IFN β-1b; similar findings are reported in the literature. As has been reported by other authors, we did detect changes related to specific taxa among patients with MS.2–6,9
At the phylum level, we found statistically significant differences between patients with MS and controls in the abundance of Firmicutes, Proteobacteria, Actinobacteria, and Lentisphaerae, of which the former 3 belong to the 4 main phyla found in the gastrointestinal tract. Similarly, Mowry et al.7 describe differences in the abundance of Firmicutes and Actinobacteria between patients with MS and controls, although they also report differences in Bacteroidetes. The discrepancies between studies regarding the abundance of Bacteroidetes may be explained by methodological differences: Mowry and colleagues included patients with vitamin D deficiency and used the PhyloChip Assay system (Second Genome, Inc.). Similarly, Miyake et al.2 report decreased abundance of Firmicutes and Bacteroidetes, although this difference was not statistically significant. The increased abundance of Actinobacteria detected in our study has previously been described in children4 and in Japanese adults,2 although the latter study did not find a statistically significant difference between patients with MS and controls. The agreement between these 3 studies suggests that the phylum Actinobacteria may play an important role in MS, similarly to its reported involvement in inflammatory bowel disease.20,21
The changes reported in patients with MS in abundance of the phyla Actinobacteria,Firmicutes, and Lentisphaerae correspond to the group of untreated patients, with no significant differences detected between controls and patients receiving IFN β-1b. Future research should study causality, assessing whether a treatment-related recovery is observed in these phyla, which would be compatible with these findings. Our results for Actinobacteria are not consistent with the findings of Tremlett et al.,4 who report that immunomodulatory treatment increases the differences observed against controls; however, that study was performed with paediatric patients, in which the microbiota is less well established, with age being a key factor in microbiota composition.22 Furthermore, as the study did not aim to find treatment-related changes, it included children being treated with various drugs (glatiramer acetate, IFN, natalizumab, and corticosteroids).
The phylum Firmicutes includes the genus Clostridum, which is involved in the production of short-chain fatty acids (particularly butyrate) and in the differentiation of T cells into regulatory T cells; this genus therefore protects against autoimmune and inflammatory diseases.1,23 Within this genus, and contrary to expectations, C. bolteae was more abundant in patients with MS, independently of whether they were receiving IFN. Also within the order Clostridiales, we observed increased abundance of Faecalibacterium sp. in patients with MS. However, no differences were found in Faecalibacterium prausnitzii, a species reported to protect against various inflammatory diseases24–27; this bacterium therefore appears not to be involved in the pathogenesis or development of MS. The remaining members of the order Clostridiales for which we did find differences belonged to Clostridium cluster XIVa, which produces butyrate and as a consequence has been associated with an anti-inflammatory environment and health promotion.23,28 This may explain the reduced abundance of this order in patients with MS as compared to controls in our sample, independently of whether patients were receiving treatment. To conclude the discussion of Firmicutes, we should address the 2 species belonging to the order Selenomonadales and the family Veillonellaceae, in which we observed a significant difference between healthy controls and untreated patients, with treated patients showing similar abundance. An interesting trend towards statistical significance was also observed in the comparison between treated and untreated patients; a different study design with a direct comparison would have shown a significant difference. The physiological role and clinical relevance of this observation should be addressed in future research.
Regarding members of the phylum Bacteroidetes, order Bacteroidales, extensive research has been conducted into the species Bacteroides fragilis for its possible protective effect against experimental autoimmune encephalomyelitis1; a statistically significant difference is reported in the abundance of this species between paediatric patients with MS and controls.4 However, our results coincide with those of Miyake et al.,2 finding no treatment- or disease-related change in the abundance of B. fragilis. Further studies should address in greater detail the role of this species in MS and the reason for the discrepancy between findings from adult and paediatric populations.
At the genus level, Prevotella was markedly less abundant in patients with MS; this is consistent with other authors’ findings.2,3,5 These data support the hypothesis of an association between MS and reduced abundance of Prevotella; reduced abundance of this genus has been shown to be involved in type 2 diabetes mellitus and non-alcoholic fatty liver disease.29,30 At the species level, our results for P. copri are of great interest, as in addition to a significant reduction in its abundance among patients with MS (which coincides with the findings of other authors2,4), we also detected significantly higher abundance among patients receiving IFN β-1b than in untreated patients. The action mechanism of IFN β-1b in MS is multifactorial, and is not fully understood. The drug appears to directly increase the expression and the concentration of anti-inflammatory agents, and downregulates proinflammatory cytokines31; as a type 1 IFN, it can interact with the intestinal epithelium, regulating the microbial ecosystem.32 Our findings raise the question of whether IFN β-1b-associated changes in microbiota composition may have anti-inflammatory effects, which could in part explain the mechanism of action by which the drug reduces MS progression and relapses.
The phylum Actinobacteria includes the genus Bifidobacterium, some of whose members are included in VSL#3, a commercial probiotic mixture with several uses33; reduced abundance of Bifidobacterium has been associated with allergies, inflammatory bowel disease, irritable bowel syndrome, colorectal cancer, and diarrhoea.34 Administration of some bacteria from this genus has been shown to yield an improvement in experimental autoimmune encephalomyelitis, mediated by IL-10 and regulatory T cells.1 Despite this, we detected greater abundance of B. longum in patients with MS than in controls. Therefore, while our results contradict the evidence from animal studies, they are consistent with findings on the role of the genus Bifidobacterium in MS published by Tremlett et al.4 (in a paediatric population) and Miyake et al.,2 although the latter study did not demonstrate a statistically significant effect. The exact role of Bifidobacterium and particularly B. longum in MS merits further study.
Finally, Archaea, a domain involved in inflammation (through production of TNF), inflammatory bowel disease,35 and obesity and the associated metabolic alterations,36 was detected at lower levels in patients than in controls. Our findings contradict those of Jangi et al.,5 who report increased abundance of Methanobrevibacteriaceae (a genus in the phylum Euryarchaeota) and increased breath methane concentration in patients with MS as compared to healthy controls. These discrepancies may be explained by differences between studies in terms of patient characteristics (especially disease duration), highlighting the need for further research.
Despite the interest of our findings, our study does present certain limitations, such as the small sample size and the lack of a consensus about the most appropriate controls to use in studies into the microbiota.4 Nevertheless, the sample size used does not differ excessively from those used in other metagenomic studies of patients with MS.2–6,9 Other limitations include the fact that we did not control for some factors impacting the microbiota, such as stress or diet. Additionally, given the study's descriptive nature, we were unable to establish causality (the classic “chicken and egg” question: whether the changes in the microbiota precede the onset of MS or vice versa) or whether IFN β-1b was responsible for the changes observed in patients receiving the drug.
However, our findings do contribute to the understanding of the microbiota in patients with MS, with this being the first study to address this issue in Spain and the Mediterranean, whose population differs greatly from those studied to date in terms of lifestyle and diet. These differences are invaluable, enabling the design of future studies into the microbiota's involvement in the pathophysiology of the disease, its potential as a prognostic marker, and different strategies for evaluating the outcomes of recovery from dysbiosis. Furthermore, the similarity between the microbiota profiles of patients receiving IFN β-1b and controls, and the differences between the profiles of treated and untreated patients, is of great interest and may lead to future studies addressing whether the improvements observed after treatment are explained, at least to some extent, by the drug's influence over the microbiota. All these questions warrant further research with future studies aiming to establish a causal relationship between microbiota alterations and MS, and the influence of treatment. This would enable exploration of the possibility of correcting dysbiosis associated with MS as a potential strategy for the treatment and prevention of the disease.
FundingThis study was funded by the neurology department of Hospital San Pedro, La Rioja (Spain) and the Rioja Salud Foundation (http://www.fundacionriojasalud.org/). Neither organisation had any involvement in study design, data collection or analysis, the decision to publish, or drafting of the manuscript.
Conflicts of interestThe authors have no conflicts of interest to declare.
We are grateful to all the participants in the study. We would also like to thank María Jesús Villanueva-Millán for her invaluable technical assistance, and Enrique Ramalle Gomara for his sage advice regarding the statistical analysis.
Please cite this article as: Castillo-Álvarez F, Pérez-Matute P, Oteo JA, Marzo-Sola ME. Composición de la microbiota intestinal en pacientes con esclerosis múltiple. Influencia del tratamiento con interferón β-1b. Neurología. 2021;36:495–503.