In order to determine the presence and genetic diversity of Chlamydia spp. in the north-eastern area of Buenos Aires province, Argentina, conjunctival, oropharyngeal, cloacal swab and tissues were collected from a total of 90 psittacine pet birds of different age and clinical manifestations. Through molecular methods, Chlamydiaceae was detected in 30% (27/90) of the samples, out of which 70.3% (19/27) were positive for Chlamydia psittaci and 14.9% (4/27) for Chlamydia abortus. Nine C. psittaci positive samples were genotyped by ompA gene sequences, 8 clustered within genotype A and 1 within genotype B. A significant association was observed between the presence of Chlamydia spp. and the manifestation of clinical signs compatible with chlamydiosis, as well as with the age of the birds (younger than one year old). This report contributes to the improvement of our understanding of chlamydial agents in our country.
Con el objetivo de determinar la presencia de Chlamydia spp. en psitácidos del área noreste de la provincia de Buenos Aires y conocer su diversidad genética, se recolectaron y analizaron mediante métodos moleculares hisopados conjuntivales, orofaríngeos, cloacales y tejidos de un total de 90 psitácidos de diferentes edades y con diversas manifestaciones clínicas. El 30% (27/90) de las muestras procesadas fueron positivas para Chlamydiaceae; el 70,3% (19/27) de estas resultaron positivas para Chlamydia psittaci y el 14,9% (4/27) para Chlamydia abortus. Nueve muestras positivas para C. psittaci fueron genotipificadas por secuenciación del gen ompA: 8 correspondieron al genotipo A y una al genotipo B. Se observó una asociación significativa entre la presencia de Chlamydia spp. y la manifestación de signos clínicos compatibles con clamidiosis, como así también con la edad de las aves (menores de un año). Este informe contribuye a mejorar nuestro conocimiento de los agentes clamidiales en nuestro país.
Chlamydia psittaci, the etiologic agent of chlamydiosis, is a highly infectious obligate intracellular bacterium spread worldwide11. C. psittaci infection has been described in several species of birds and mammals, and is recognized as one of the main zoonotic diseases transmitted by birds15. The severity of avian chlamydiosis varies depending on the species and age of the infected bird, as well as on the genotype of chlamydia involved. It may present itself as an inapparent infection or exhibit various clinical signs that can lead to severe illness with a high rate of mortality11.
C. psittaci has been initially classified into 9 genotypes: A–F, E/B, M56, and WC, using the sequence analysis of the outer membrane protein A (ompA)11. Each genotype exhibits strong host preference: A in Psittacine birds, B in Columbiformes; genotype C was detected in Anseriform and Galliform birds, D in Galliformes, E in Columbiform birds, Anseriformes and other bird species, F in Psittacine and Galliform birds, WC in cattle and M56 in rodents. Subsequently, eight new genotypes were proposed (1V, 6N, Mat116, R54, YP84, CPX0308, I and J), having been found in psittacine and wild birds. All genotypes have zoonotic potential which makes them a risk for human health11,13.
In humans, symptoms are generally wide-ranging and influenza-like. However, occasionally they may include severe pneumonia, endocarditis, encephalitis and renal disease. Infection occurs through the inhalation of aerosolized droppings and secretions while handling animals, carcasses or infected tissues, where feces and feathers play a key role in zoonotic transmission1. People frequently exposed to or in contact with pet or captive birds, either in their free time or occupationally, are the ones who are at a higher risk of becoming infected1,6,15.
Chlamydia abortus causes abortions in ruminants and is one of the most common causes of abortions in ovines and caprines. This bacterium can also infect bovines, swine, horses and deer. C. abortus constitutes a zoonotic risk for humans because it can lead to an influenza-like illness, or even, on rare occasions to pneumonia. Infection in pregnant women can result in abortion and serious complications such as acute kidney failure, disseminated intravascular coagulation (DIC) or respiratory failure requiring mechanical ventilation4. In the last ten years, the avian C. abortus strain has been detected in various species of wild birds, although until now the role it plays in causing avian diseases and the zoonotic transmission of these avian strains is unclear8,13.
Despite the zoonotic importance of C. psittaci and C. abortus, only few analyses have been conducted to support evolutionary and epidemiological investigations. Therefore, the aim of this study was to detect the presence and begin with the characterization of the genetic diversity of Chlamydia through molecular methods in psittacine pet birds received in the Cátedra de Patología de Aves y Pilíferos at Facultad de Ciencias Veterinarias, Universidad de La Plata, for diagnosis and/or clinical control.
Between March 2013 and March 2015, swab samples and organs from 90 psittacine pet birds coming from the northeastern region of Buenos Aires province, Argentina, were included. Samples were taken from 57 specimens of Amazona aestiva, 29 of Myiopsitta monachus, 1 Cyanoliseus patagonus, 1 Nymphicus hollandicus, 1 Melopsittacus undulatus and 1 Agapornis sp. The conjunctival, oropharyngeal and cloacal swab samples (n=81) were respectively obtained for each bird, using a single swab. The samples were chilled and submitted to the laboratory. We also studied the visceral organs belonging to other necropsied birds (n=9). The organ pools (liver, lung, air sacs and spleen) were processed, placed in sterile tubes and stored at −20°C until required for analysis. Birds of different ages with symptomatology and lesions compatible with chlamydiosis, as well as those showing no evident manifestations of the disease, were analyzed.
Each swab was resuspended in 600μl of phosphate-buffered saline (PBS) and homogenized by vortex. The pellet was centrifuged and resuspended in 200μl and then Quick-gDNA™ MiniPrep Zymo Research USA was used to extract DNA, according to the manufacturer's instructions.
Approximately 25μg of the organs from the necropsied birds were used for DNA extraction using PureLink™ Genomic DNA Mini Kit Invitrogen USA, according to the manufacturer's instructions. Template DNA was stored at −20°C until required for analysis.
Three microliters of DNA were used in a real-time polymerase chain reaction, specific for the Chlamydiaceae family and target 23S rRNA, as described by Ehricht et al.3 Positive samples underwent another real-time PCR, specific for C. psittaci ompA gene, using CppsOMP1-F 5′-CACTATGTGGGAAGGTGCTTCA-3′ and CppsOMP1-R 5′-CTGCGCGGATGCTAATGG-3′ primers8. The final PCR volume of the reaction was 25μl, containing 12.5μl of Bio-RadiQ™ SYBR GREEN Supermix®, 3μl of DNA sample, 1μl (0.2μM) of each primer and 7.5μl of distilled water. Amplification was carried out under the following protocol: initial denaturation at 95°C for 2min, followed by 45 cycles of denaturation at 95°C for 5s, annealing at 60°C for 15s and extension at 72°C for 30s, then final extension for 2min at 72°C. The dissociation curve was done with 51 cycles at 70°C for 30s, with a change in temperature of 0.5°C and a final temperature of 95°C.
Chlamydia-positive samples by real-time PCR underwent a Nested-PCR to amplify fragments of variable domains III and IV of the Chlamydia spp. ompA gene (576 pb) in the first run, and for C. psittaci (404 pb), C. pecorum (441 pb) and Chlamydia spp. (450 pb) in the second run10. All PCR reactions were performed using IQTM5 Multicolor Real-Time PCR Detection System (BIO-RAD Laboratories) equipment.
The products obtained were purified with AccuPrep® Gel Purification Kit (Bioneer Corporation, Seoul, Korea) and sent for direct nucleotide sequencing reaction using internal primers to Macrogen, Inc. (Seoul, Korea).
The resulting sequences were aligned and trimmed using Clustal X (Conway Institute UCD Dublin, Dublin, Ireland) and the consensus sequences were compared through BLASTn 2.2.19 to other Chlamydia ompA gene fragments obtained from the GenBank data bank. The dendrogram was constructed using the Tree Explorer module of the MEGA7 program with the Neighbor-Joining method and the p-distance parameter. Statistical support was calculated through non-parametric bootstrapping with 1000 pseudo-replications14.
The association analysis between the presence of Chlamydia spp. and the following variables: the clinical signs (the presence or absence of clinical signs), age (birds younger or older than one year of age), clinical signs and age (sick birds younger than one year of age, or older than one year of age) was performed through odds ratio estimation (OR) (IC95%).
The statistical analysis was performed using the Epidemiological Analysis from the Tabulated Data program, Epidat 3.1 (Dirección Xeral de Saúde Pública, Consellería de Sanidade-Xunta de Galicia, Área de Análisis y Sistemas de información Sanitaria, Organización Panamericana de la Salud). For all the analyses performed, the statistical significance was determined as p<0.05.
Overall, out of all the Psittaciform bird samples screened, 96.7% (87/90) of the birds were native to Argentina, whilst 3.3% (3/90) were exotic. Likewise, 52.2% (47/90) of the samples belonged to birds younger than one year of age, while 47.8% (43/90) belonged to those older than one year of age. Additionally, 54.4% (49/90) of the samples belonged to birds whose symptomatology or macroscopic lesions were compatible with chlamydiosis, and 45.6% (41/90) to clinically normal birds.
Out of the 90 specimens, 30% (27/90) of the samples were found to be positive by real time PCR assay (23S rRNA), 48.1% (13/27) of them belonged to the A. aestiva species, 44.4% (12/27) to M. monachus, 3.7% (1/27) to C. patagonus, and 3.7% (1/27) belonged to N. hollandicus.
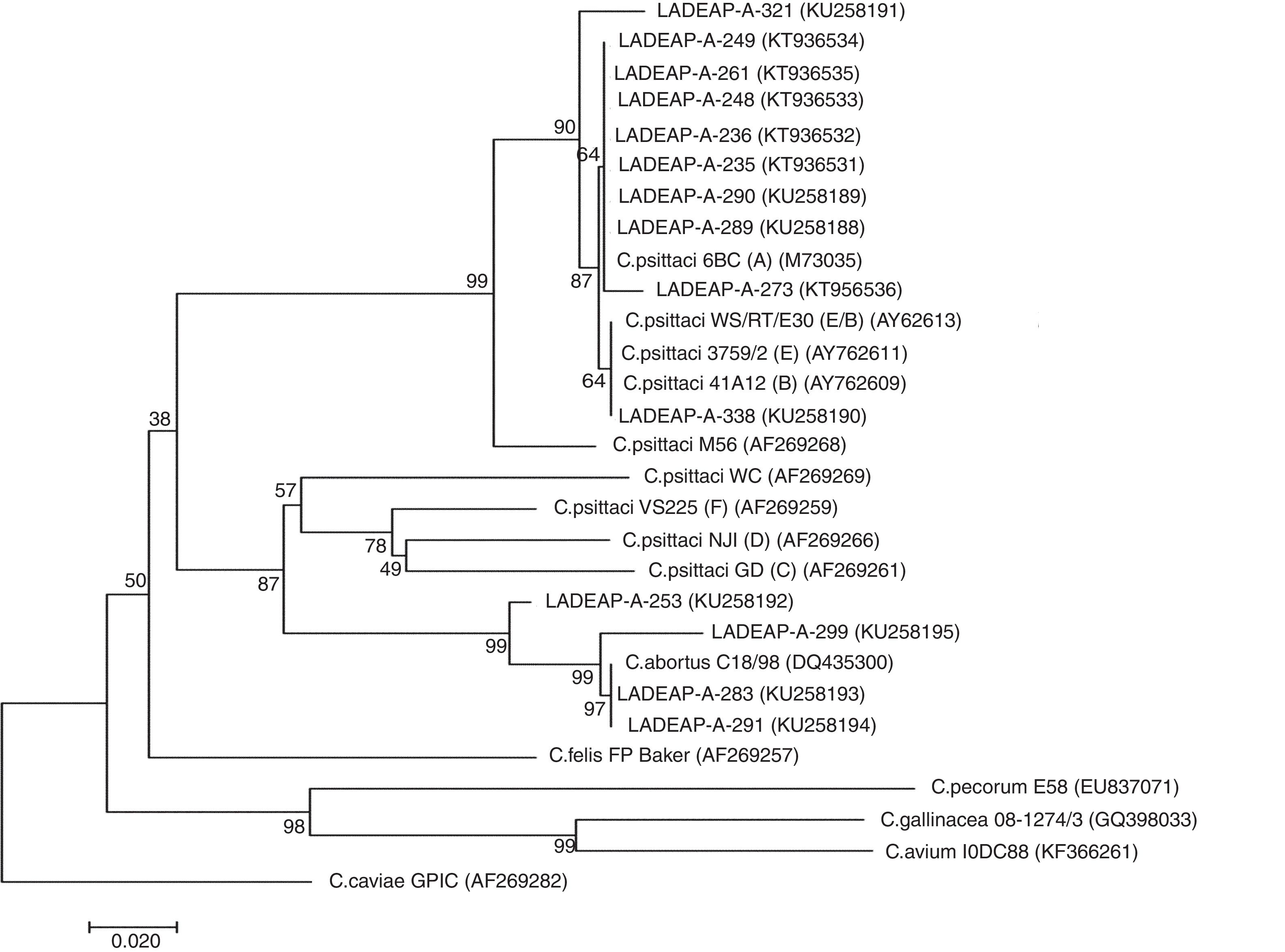
In addition, of the 27 Chlamydia-positive samples, 24 were swabs and 3 belonged to others necropsied birds, with 70.3% (19/27) of the samples analyzed found to be C. psittaci-positive, while 14.9% (4/27) tested positive for C. abortus (Table 1) (Supplementary material). With regard to the products obtained, 51.2% (14/27) of them were sequenced and genetically characterized because they showed good quality and DNA concentration. The analysis of the ompA gene sequence dendrogram for C. psittaci revealed that 8 samples (LADEAP-A-235, LADEAP-A-236, LADEAP-A-248, LADEAP-A-249, LADEAP-A-261, LADEAP-A-273, LADEAP-A-289, LADEAP-A-290) clustered with C. psittaci genotype A. One sample (LADEAP-A-338) clustered with C. psittaci genotype B, while the sequence of sample LADEAP-A-321 clustered differently to the rest of the C. psittaci genotypes. Samples LADEAP-A-253 (A. aestiva), LADEAP-A-283 (M. monachus), LADEAP-A-291 (M. monachus), and LADEAP-A-299 (A. aestiva) clustered with C. abortus sequences (Fig. 1).
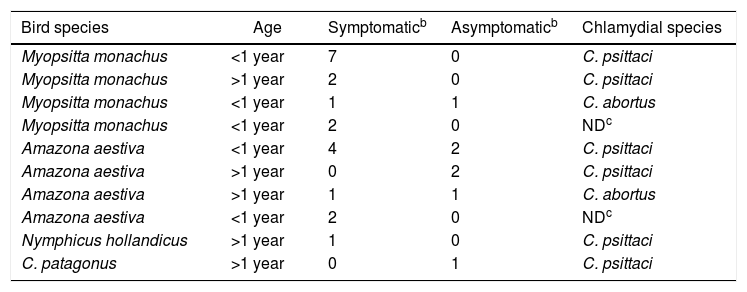
Characteristics of Chlamydia spp. positive casesa
| Bird species | Age | Symptomaticb | Asymptomaticb | Chlamydial species |
|---|---|---|---|---|
| Myopsitta monachus | <1 year | 7 | 0 | C. psittaci |
| Myopsitta monachus | >1 year | 2 | 0 | C. psittaci |
| Myopsitta monachus | <1 year | 1 | 1 | C. abortus |
| Myopsitta monachus | <1 year | 2 | 0 | NDc |
| Amazona aestiva | <1 year | 4 | 2 | C. psittaci |
| Amazona aestiva | >1 year | 0 | 2 | C. psittaci |
| Amazona aestiva | >1 year | 1 | 1 | C. abortus |
| Amazona aestiva | <1 year | 2 | 0 | NDc |
| Nymphicus hollandicus | >1 year | 1 | 0 | C. psittaci |
| C. patagonus | >1 year | 0 | 1 | C. psittaci |
Neighbor-joining dendrogram based on the comparison of the ompA gene fragment (about 380bp) of Chlamydia. Samples that belong to this study have the prefix LADEAP-A- and GenBank accession numbers provided. Numbers above branches are bootstrap values as a percentage of 1000 pseudo replicates. C. caviae GPIC was used as an outgroup. The scale bar shows the sequence diversity percentage.
C. pecorum DNA was not detected in any of the samples tested.
In 74.07% (20/27) of Chlamydia-positive bird samples, the existence of clinical signs compatible with disease were clearly observed. A significant association between the presence of Chlamydia spp. and the manifestation of clinical signs compatible with chlamydiosis was detected. A higher chance of finding the agent in sick birds younger than one year of age was also observed (Table 2).
OR and IC95% values estimated for each category
C. psittaci is one of the most studied avian bacteria, with psittacine birds as one of the main sources of infection for humans1,15. Psittacosis is a widely spread disease which has been known for decades in Argentina, with 638 reported cases and 82 confirmed cases in humans for the 2013–2014 biennium7. These data may not reflect the reality of psittacosis in our country, as it is frequently subdiagnosed. Patients’ history of exposure to or contact with birds or other animals is often omitted in the medical consultation. On the other hand, the prevalence in psittacine pet is unknown. Throughout this study, the use of molecular methods allowed us to detect and genetically characterize the members of the Chlamydiaceae family present in psittacine pet birds. Through the use of these strategies, the examined samples revealed the existence of C. psittaci and C. abortus in psittacine birds with clinical symptomatology of the disease, as well as in other clinically normal birds.
Chlamydiaceae DNA was detected in 30% of the tested birds, a result which is higher than the one obtained by other authors in our country who found a 17.4% of DNA5 and higher even than that reported in Brazil (3.4–10.6%)12, though in this case the study was focused only on C. psittaci. In a recent study carried out in the center of the country, C. pneumonie, C. pecorum and the mixed infection of C. pneumonie/C. pecorum were found in caged psittacine birds, kept as pets in households and caged in zoos5. In contrast to what has been reported in previous studies on the subject, we have clearly detected C. psittaci and C. abortus in psittacine pet birds.
C. psittaci was the Chlamydia species most often detected in the analyzed bird samples, which reflects its circulation among psittacine pet birds in the area where the study was conducted. One brief report from Argentina showed that C. psittaci from three parrots related to human cases had the same genotype2.
Our report showed that 7.7% of the asymptomatic birds tested positive for Chlamydiaceae. In sick birds, a significantly higher chance was found in those younger than one year of age. Due to the results obtained, it would seem prudent to carry out diagnostic tests routinely to detect the presence of C. psittaci in all recently-acquired psittacine birds, and to re-evaluate these birds periodically throughout their lives. Likewise, 22.2% of birds harboring C. psittaci showed chlamydiosis-compatible symptomatology, a percentage that is in line with reports by Varompay et al.15 in Belgium. These authors have reported that clinically healthy birds that tested negative for Chlamydia had previously received treatments with antibiotics for Chlamydia. In our study, the birds had not received any antibiotic treatment prior to sample acquisition. Our study reveals an important circulation of C. psittaci genotype A in psittacine pet birds within the area where the study was carried out, in line with what has been previously found in other countries9. Recently in Argentina, Cadario et al.2 found only C. psittaci genotype A in human cases of psittacosis and related birds in some regions of Argentina. However, in the central region of our country, Frutos et al.5 found genotype WC present both in humans and birds. Genotype A is considered to be one of the most virulent strains in birds and humans, being psittacine birds the primary carriers of this genotype, and it has been regarded as the main source of infection for humans6,11. Another genotype was detected in the present study, genotype B, which is mainly related to pigeons, although it has been reported in psittacines as well9. Kalmar et al.6 found C. psittaci genotype B in 3 workers from a bird refuge who were in contact with birds in which the same genotype was detected. To our knowledge, this is the first report of genotype B in birds in Argentina, which shows a new genotype circulating in birds from our country.
Given the insufficient DNA obtained in those Chlamydiaceae-positive samples by real time PCR, it was not possible to type them and, consequently, it was not possible to determine if other members of the Chlamydiaceae family were present in the studied birds.
In the present study, we found C. abortus in 4 psittacine birds, 2 of which showed clinical signs compatible with chlamydiosis while the remaining 2 were asymptomatic. These asymptomatic carriers may represent a reservoir of C. abortus in our area.
However, the neighbor-joining dendogram used to assign chlamydial species is built based on a 380bp fragment, which is too short to draw definite conclusions. A larger fragment and a couple of different loci should be analyzed in order to obtain more accurate results13.
As far as we know, there are no previous reports of C. abortus in birds in Argentina. On the other hand, it provides further proof of its circulation in pet birds in our region; nevertheless, the epidemiological aspects and the importance of C. abortus in these birds are yet to be established. Based on these findings, it should be kept in mind that keeping psittacine birds in captivity implies the zoonotic risk of C. psittaci infections and the potential risk of C. abortus infections.
In those analyzed birds which tested positive for Chlamydia, the manifestations of clinical signs may have depended on the virulence of the strain, host factors and/or the various host-pathogen interactions6.
A. aestiva and M. monachus were the most studied birds and also, the most sought after as pets in Argentina, commonly due to illegal trafficking. Frequently these birds are kept until they are sold in inadequate environmental conditions, below-standard levels of hygiene and poor nutrition. Although some authors indicate that the above mentioned factors listed by us could influence the development of the disease, we cannot affirm this fact in our study because we used the convenience sampling method rather than the random sampling method with a control population.
In the literature, the studies conducted regarding Chlamydia infections on birds, have usually been restricted to the search of C. psittaci; therefore, little is known about the presence of other chlamydial agents. New findings confirm that our knowledge on the varieties of Chlamydia in birds remains partial.
This study provided further identification and molecular characterization of Chlamydia in Argentina psittacine pet birds, and it represents a contribution to the improvement of our understanding of the various bacteria present in the animal kingdom.
Conflict of interestThe authors declare that they have no conflicts of interest.
The authors thank Nancy Arias and Maria Cecilia Netri for assistance in DNA extraction and Maira Poggio for her help in English traslation of manuscript. This study was supported in part by Programa de Incentivo a Docentes e Investigadores de la UNLP (11/V219) and PICTO-ANLIS (0180-2011).