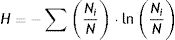Diversity and abundance of the denitrifying genes nirK, nirS and nosZ were investigated in cow manure compost using polymerase chain reaction–denaturing gradient gel electrophoresis (PCR–DGGE) and real-time quantitative PCR (qPCR), respectively. These three genes were detected in all the stages of the composting process. Phylogenetic analysis showed that the nirK gene was closely related to Rhizobiales, Burkholderiales, the nirS gene was closely related to Pseudomonadales and Burkholderiales, and the nosZ gene was closely related to Rhodospirillales, Rhizobiales, Pseudomonadales, and Alteromonadales. qPCR results showed that the abundance of these three genes (nirK, nirS and nosZ) reached the peak value in the late thermophilic stage of composting and abundance of the nirK gene was higher than that of the nosZ gene and the nirS gene. Redundancy analysis (RDA) showed that the diversity of the nirK and nirS genes was significantly correlated with ammonium (p<0.05), the diversity of the nosZ gene was significantly correlated with pH (p<0.05) and the abundance of the nirK nirS and nosZ genes was significantly correlated with temperature (p<0.05).
La diversidad y la abundancia de los genes desnitrificadores nirK, nirS, nosZ en el compost de estiércol de vaca se investigaron por medio de la reacción en cadena de la polimerasa seguida de electroforesis en gel con gradiente de desnaturalización (PCR-DGGE) y por PCR cuantitativa (qPCR) en tiempo real, respectivamente. Estos 3 genes fueron detectados durante todas las fases del compostaje. El análisis filogenético mostró estrecha relación del gen nirK con Rhizobiales y Burkholderiales, del gen nirS con Pseudomonadales y Burkholderiales y del gen nosZ con Rhodospirillales, Rhizobiales, Pseudomonadales y Alteromonadales. Los resultados de la qPCR mostraron que la abundancia de los genes nirK, nirS y nosZ alcanzó el valor máximo en la fase termofílica tardía del compostaje, y que la abundancia del gen nirK era más elevada que los de los genes nosZ y nirS. El análisis de redundancia (RDA) mostró que la diversidad de los genes nirK y nirS estaba significativamente correlacionada con la concentración de amonio (p<0,05), mientras que la del gen nosZ estaba significativamente correlacionada con el pH (p<0,05). También mostró que la abundancia de los genes nirK, nirS y nosZ estaba significativamente correlacionada con la temperatura (p<0,05).
As a well-known traditional method to stabilize livestock manure, the composting process can convert biodegradable components into nuisance free and sanitary materials by microbial communities under controlled conditions16,33,36–38. The final product can be reused as organic fertilizer for crop production. Therefore, composting plays a very important role in the circulation of resources and environmental protection.
During composting, the organic nitrogen contained within initial fresh manure is degraded into ammonia-N(NH4+–N) by a wide variety of microorganisms6. In the presence of oxygen, NH4+ is sequentially oxidized to NO3– by specific groups of bacteria. However, in the absence of oxygen, NO3– can be converted into gaseous products such as nitric oxide (NO), nitrous oxide (N2O), or N2 by various microbes as a respiratory electron acceptor. This process is known as denitrification, and can cause lower fertilizer values in the compost product and higher levels of nitrous oxide (N2O)3. Nitrous oxide, which is produced as an intermediate of denitrification, has a noteworthy global warming potential by absorbing infrared radiation34.
During the denitrification process, nitrate is reduced by nitrate reductases, nitrite reductases, nitric oxide reductases and nitrous oxide reductases, which are encoded by the napA gene or the narG, nirS or nirK, nor and nosZ genes respectively40. A variety of samples have been studied using the nirK, nirS and nosZ genes as molecular markers to indicate the diversity and abundance of denitrifiers in various environments7,8,23,30,31. It is known that microorganisms are affected by environmental factors and the community of denitrifiers is no exception. A large number of studies in the literature have reported that the abundance and community composition of the functional genes involved in denitrification can be affected by changes in environmental factors12,13,20. However, few efforts have been made to examine the effects of environmental factors on diversity and quantification of denitrifiers during composting, especially in animal manure compost, which is of significance to understanding the process of nitrogen transformation during composting and management of animal manure composting.
In the present study, the diversity and abundance of denitrifies were assessed by targeting the nirK, nirS and nosZ genes using DGGE and qPCR respectively. Moreover, the relationship between diversity and abundance of the denitrifying genes and physico-chemical parameters were investigated.
Materials and methodsComposting set-up and sample collectionComposting of cow manure and rice straw was conducted in a tank (0.5m×0.5m×1.0m) with forced aeration for 36 days. C/N ratio of the mixture of cow manure and rice straw was about 30:1 and the moisture content was 65%. The sub-samples were collected from three depths (20cm, 40cm, 60cm from the top) of the composting pile on days 1, 2, 3, 4, 7, 11, 14, 19, 25, and 36. The sub-samples were mixed and then stored immediately under −80°C and 4°C.
Physical and chemical determinationsTemperature in the middle of the composting pile was measured using digital thermometers. pH was determined with a pH meter after shaking the fresh samples in water at a ratio of 1:10 (w/v, sample:water) at 120 rev/min for 60min. Ammonium (NH4+) and nitrate (NO3−) were extracted with 2mol/l KCl and analyzed by dual-channel flow analyzer (AA3, Germany).
DNA extraction and purificationTotal genomic DNA was extracted from compost samples according to the method described by Liu et al.22 The extracts were purified using Omega DNA Purification Kit (Omega, USA) according to instructions. The purified DNA was stored at −20°C. DNA quality was checked by electrophoresis in 1.0% agarose gel and concentration was determined by Nano-300 spectrophotometer (China).
PCR amplification and DGGE analysisFor the denaturing gradient gel electrophoresis (DGGE) analysis, PCR amplification was performed in a 25.0μl reaction mixture including 2.5μl of 10× PCR Buffer, 2.5μl of dNTP (10μmol/l), 0.5μl of DNA template, 0.2μl of forward primer and reverse primer (100μmol/l) respectively, 0.3μl of Taq DNA Super polymerase and 18.8μl ddH2O. Primer sets F1aCu (5′-ATCATGGTSCTGCCGCG-3′):R3Cu (5′-GCCTCGATCAGR TTGTGGTT-3′)22; cd3af (5′-GTSAACGTSAAGGARACSGG-3′):R3cd (5′-GASTTCGGRTGSGTCTTGA-3′)39 and nosZ-F (5′-CGYTGTTCMTCGACAGCCAG-3′):nosZ1662R(5′CGSACCTTSTTGCCSTYGCG-3′)18,32 were used for the amplification of the nirK, nirS and nosZ genes, respectively. A GC clamp (5′CGCCCGCCGCGCGCGGCGGGCGGGGCGGGGGCACGGGGGG-3′) was attached to the forward primers F1aCu, cd3af and nosZ1622R to prevent complete separation of the DNA strands during DGGE25. The PCR program was as follows: for the nirS gene, 95°C for 5min, followed by 35 cycles of 95°C for 30s, 57°C for 60s, 72°C for 50s followed by 72°C for 10min; for the nirK gene, 95°C for 5min, followed by 35 cycles of 95°C for 30s, 58°C for 60s, 72°C for 50s followed by 72°C for 10min; and for the nosZ gene, 94°C for 4min, followed by 35 cycles of 94°C for 45s, 61°C for 45s, 72°C for 45s, followed by 72°C for 7min.
DGGE analysis of the amplified nirS, nirK and nosZ genes was performed using the Dcode™ Universal Mutation Detection System (Bio-Rad, USA) according to the manufacturer's instructions. About 30.0μl of PCR products was loaded onto the 1-mm-thick 8% (w/v) polyacrylamide gel with a denaturing gradient of 40–70%. Electrophoresis was performed in 1× TAE buffer at 60°C for 12h at 120V32. The gel was stained before transillumination, the DNA bands were excised for PCR sequencing using primer sets without the GC-clamp. PCR products were purified before the cloning procedure. The gels were run in duplicate to ensure reproducibility of obtained patterns.
Real-time PCR assayAbundance of the nirK, nirS and nosZ genes was determined in triplicate using CFX-96TM Real-Time PCR Systems. The 25.0μl reaction mixtures contained 12.5μl SYBR® Premix Ex Taq II (Tli RNaseH Plus), 1.0μl PCR forward primer, 1.0μl PCR reverse primer, 2.0μl DNA template and ddH2O to complete the 25.0μl volume. The cycling condition was 95°C for 30s, followed by 40 cycles of 95°C for 5s, 60°C for 30s for the nirK, nirS and nosZ genes. The primer sets for the nirK, nirS and nosZ genes were the same as those used in the DGGE analysis.
Standard curves were obtained by tenfold serial dilutions of linearized plasmids containing the nirK, nirS and nosZ genes. Standard curves for the assays were generated by plotting the threshold cycle values versus log10 of the gene copy numbers. The amplification efficiency and linearity (R2) of standard curves for the nirK, nirS and nosZ genes were 104.5% and 0.986, 90.7% and 0.816, 100.8% and 0.904, respectively.
Sequencing and phylogenetic analysisThe PCR products of the DGGE bands were purified and cloned before sequencing (The Beijing Genomics Institute). The obtained nucleotide sequences were compared with available sequences in the GenBank database using the NCBI BLASTn program (http://www.ncbi.nlm.nih.gov/blast). Matching sequences were retrieved from NCBI. Phylogenetic trees were constructed using the neighbor-joining algorithm by MEGA428.
Statistical analysisThe data of physico-chemical parameters were statistically analyzed using the SPSS software (version 21). DGGE band position and intensity data for each sample were calculated and exported to an Excel spreadsheet prior to further statistical analysis. Band numbers and relative intensity (within lane) were quantified with the Quantity One software (version 2.0, Bio-Rad, Hercules, CA, USA) as described previously38. The Shannon diversity index (H) was calculated as:
In this formula, i is the number of bands in each DGGE profile, Ni is the relative intensity of a specific band i and N is the sum of intensities of all bands. Bands with relative intensity below 1% were discarded before the analysis. Canoco (version 4.5) was used for further determining the relationships between diversity, abundance of the nirK, nirS and nosZ genes and the physico-chemical parameters. Statistical significance was kept at p<0.05.
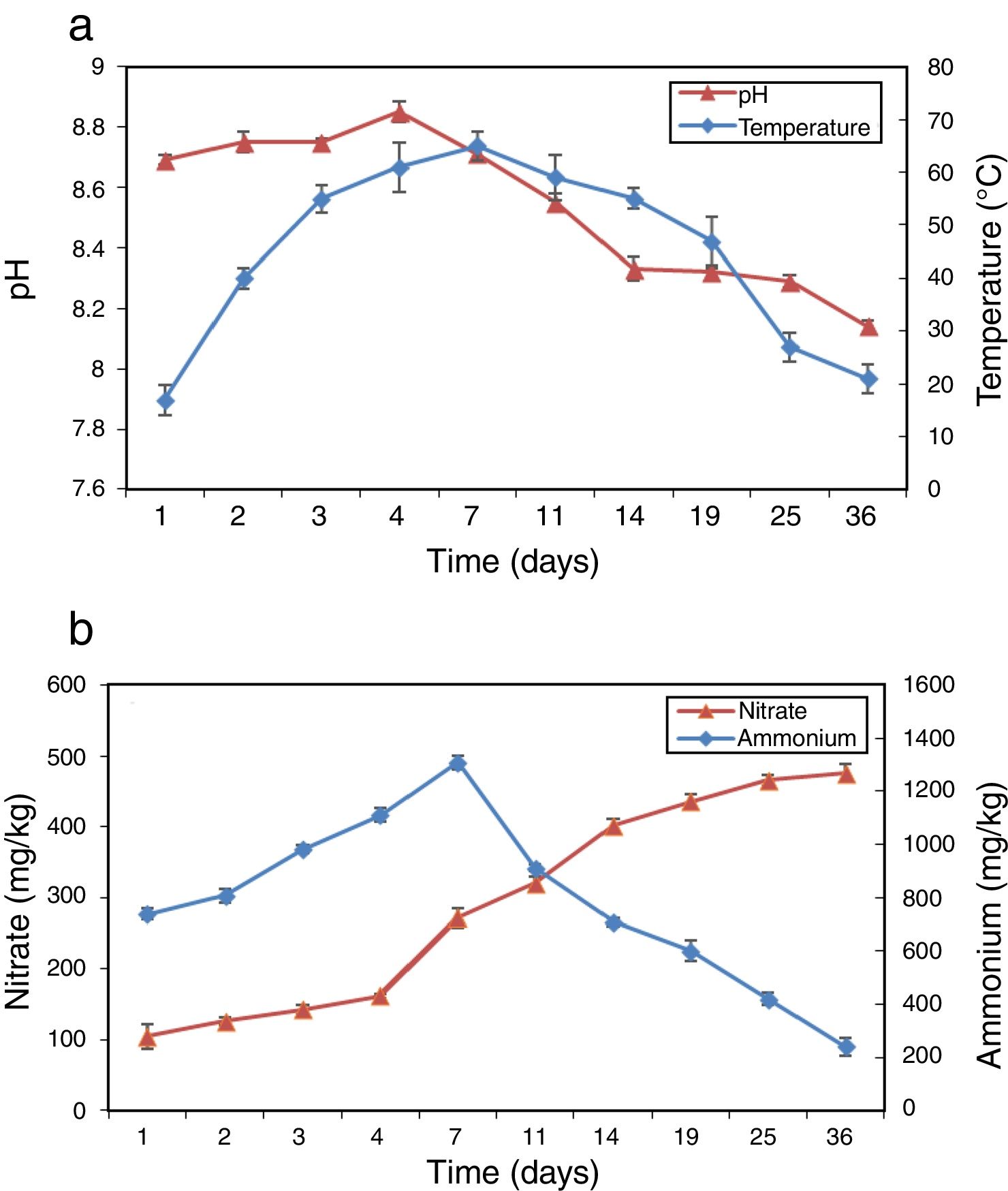
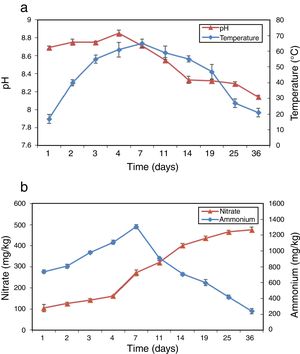
ResultsPhysico-chemical parametersThe variation of physico-chemical parameters is presented in Figure 1. During the mesophilic stage (day 1–2) the temperature rapidly reached 40°C in the compost, and then reached the peak value of 65°C on day 7 during the thermophilic stage (day 3–14). The temperature dropped from 55°C to 21°C during the cooling stage (day 15–25) and the maturation stage (day 26–36). pH increased from 8.69 to 8.85 during the first 4 days and then dropped to 8.14 at the end of composting (Fig. 1a). Figure 1b shows the profiles of NH4+–N and NO3−–N during composting. As a result of the mineralization of the organic nitrogen compounds NH4+–N accumulated rapidly in the beginning and reached peak value on day 7. Afterwards, NH4+–N began to decrease gradually until the end of composting with a level of 240.7mg/kg. NO3−–N increased continually from 105.0 to 475.2mg/kg during the whole composting process.
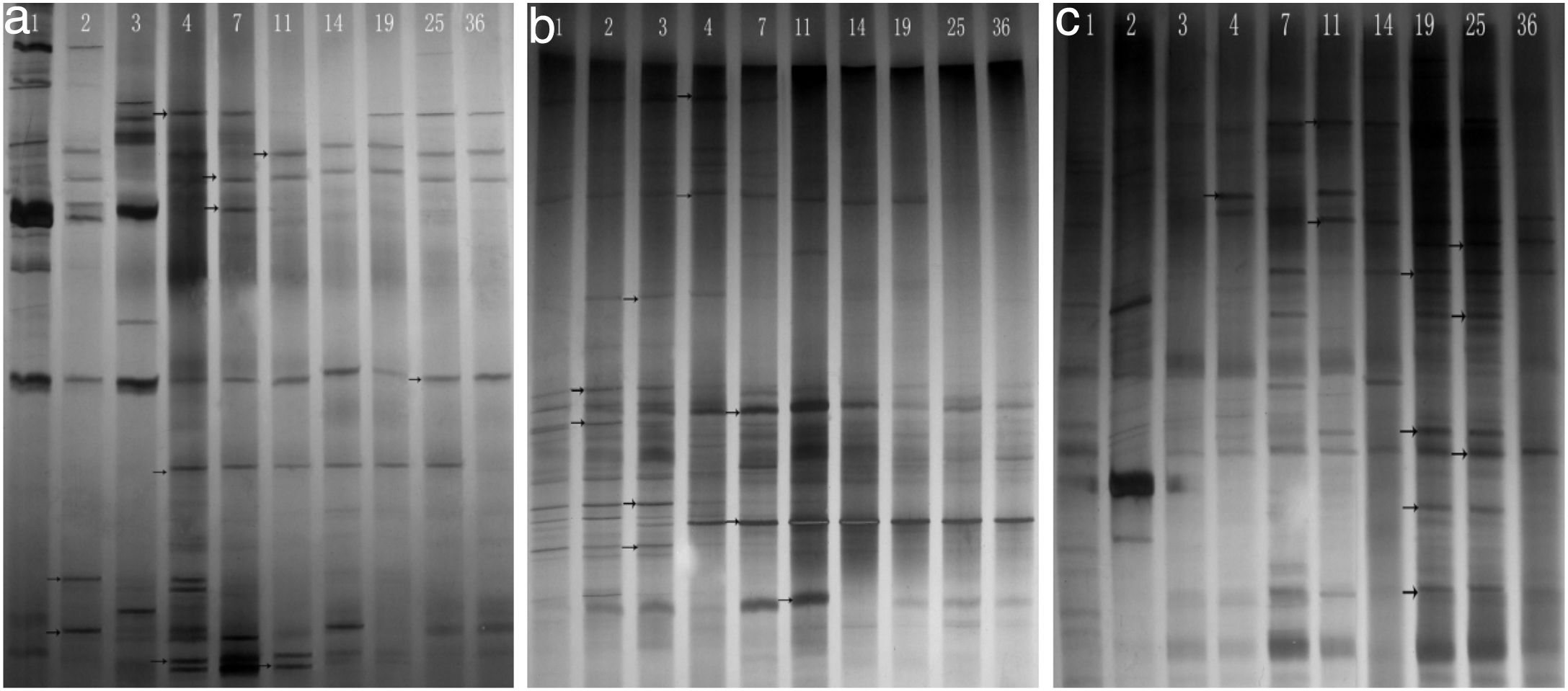
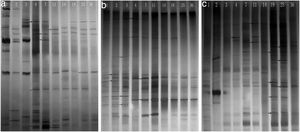
Phylogenetic diversity of the nirK, nirS and nosZ genesFingerprinting of the microbial community structure of the nirK, nirS and nosZ genes was analyzed by DGGE, in which the variations of the denitrifying species were observed (Fig. 2). DGGE profiles with two replicates for each compost sample indicated good reproducibility. High diversities were shown in the DGGE profiles of the nirK, nirS and nosZ genes. There were obvious variations between samples from different composting stages for the three genes.
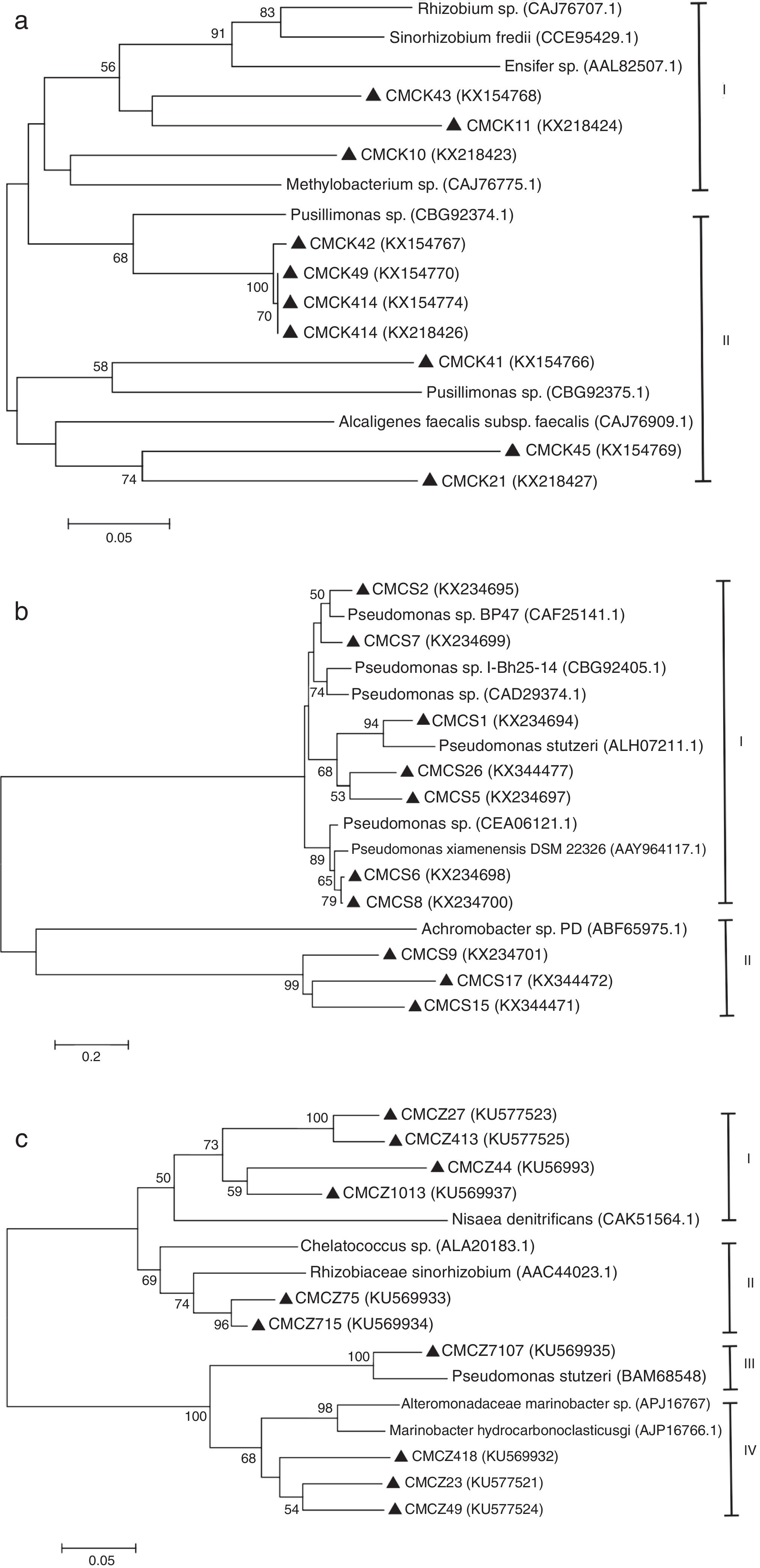
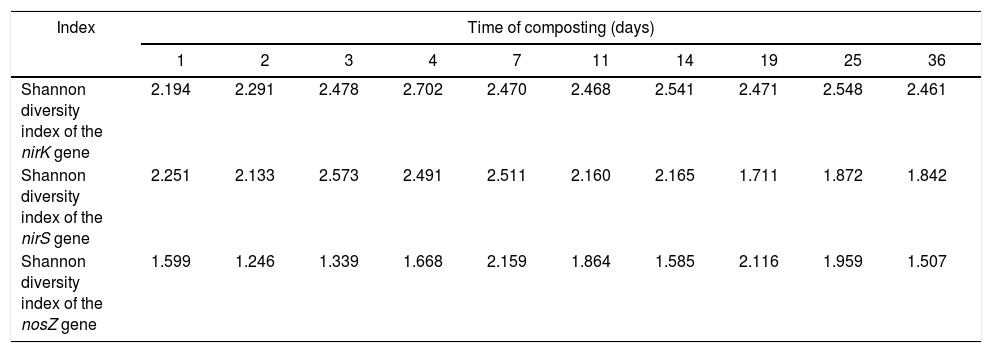
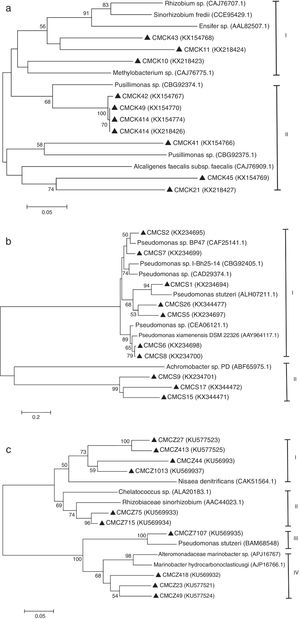
The diversity of the denitrifying community was evaluated by the Shannon diversity index. The diversity indices of the three genes were various in the different sampling times. Higher diversity indices of the nirK, nirS and nosZ genes were observed during the thermophilic stage and the Shannon values of the nirK and nirS genes were higher than those of the nosZ gene during composting (Table 1). The neighbor-joining phylogenetic trees based on the protein sequence of the three genes are shown in Figure 3. The phylogenetic tree of the nirK gene was divided into two clusters (cluster I–II). The genes belonging to cluster I and cluster II were closely related to Rhizobiales and Burkholderiales respectively. The phylogenetic tree of the nirS gene was also divided into two clusters (cluster I–II). The nirS genes in cluster I and cluster II were closely related to Pseudomonadales and Burkholderiales respectively. The phylogenetic tree of the nosZ gene was divided into four clusters (cluster I–IV). The genes in cluster I and cluster II were closely related to Rhodospirillales and Rhizobiales respectively. The genes in cluster III and cluster IV were closely related to Pseudomonadales and Alteromonadales respectively.
Shannon diversity index of DGGE profiles for each compost sample.
| Index | Time of composting (days) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 7 | 11 | 14 | 19 | 25 | 36 | |
| Shannon diversity index of the nirK gene | 2.194 | 2.291 | 2.478 | 2.702 | 2.470 | 2.468 | 2.541 | 2.471 | 2.548 | 2.461 |
| Shannon diversity index of the nirS gene | 2.251 | 2.133 | 2.573 | 2.491 | 2.511 | 2.160 | 2.165 | 1.711 | 1.872 | 1.842 |
| Shannon diversity index of the nosZ gene | 1.599 | 1.246 | 1.339 | 1.668 | 2.159 | 1.864 | 1.585 | 2.116 | 1.959 | 1.507 |
Phylogram for nirK gene (a), nirS gene (b) and nosZ gene (c) based on partial gene fragments. Bootstrap values (%) were generated from 1000 replicates, and the values >50% are shown. CMC is an acronym for cow manure compost. GenBank accession numbers are included for reference sequences used.
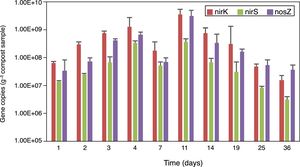
Real-time PCR assays were used to determine the copy numbers of nirK, nirS and nosZ genes in the compost samples. The abundance of nirK, nirS and nosZ genes are shown in Figure 4. The copy numbers of these three genes increased in the first 4 days of composting, however, they dropped on day 7, reached the peak on the 11th day and then declined until the end of the composting process. On the whole, the abundance of the nirK gene was higher than that of the nosZ and nirS genes in the samples collected from all the stages of composting.
During the early stage of composting rapid growth of microorganisms consumed the oxygen in the compost, causing an anaerobic environment which was favorable for the growth of denitrifiers. In addition, the abundant organic compounds in the early stage of composting may increase the number and activity of heterotrophic population, which probably carries denitrifying genes. This may explain the increase in denitrifying genes on day 4 when the temperature was rising in the compost. However, on day 7 when the temperature reached the maximum (65°C), the condition was harmful for denitrifiers survival, contributing to the decline of their abundance. A higher level of NO3−–N, which is substrate for bacteria, may be favorable for the growth of denitrifiers. Therefore, the maximum abundance of denitrifiers on day 11 could be caused by the increased level of NO3−–N (320.3mg/kg) and favorable conditions including relatively temperature (59°C) and probably a lower level of oxygen in the compost.
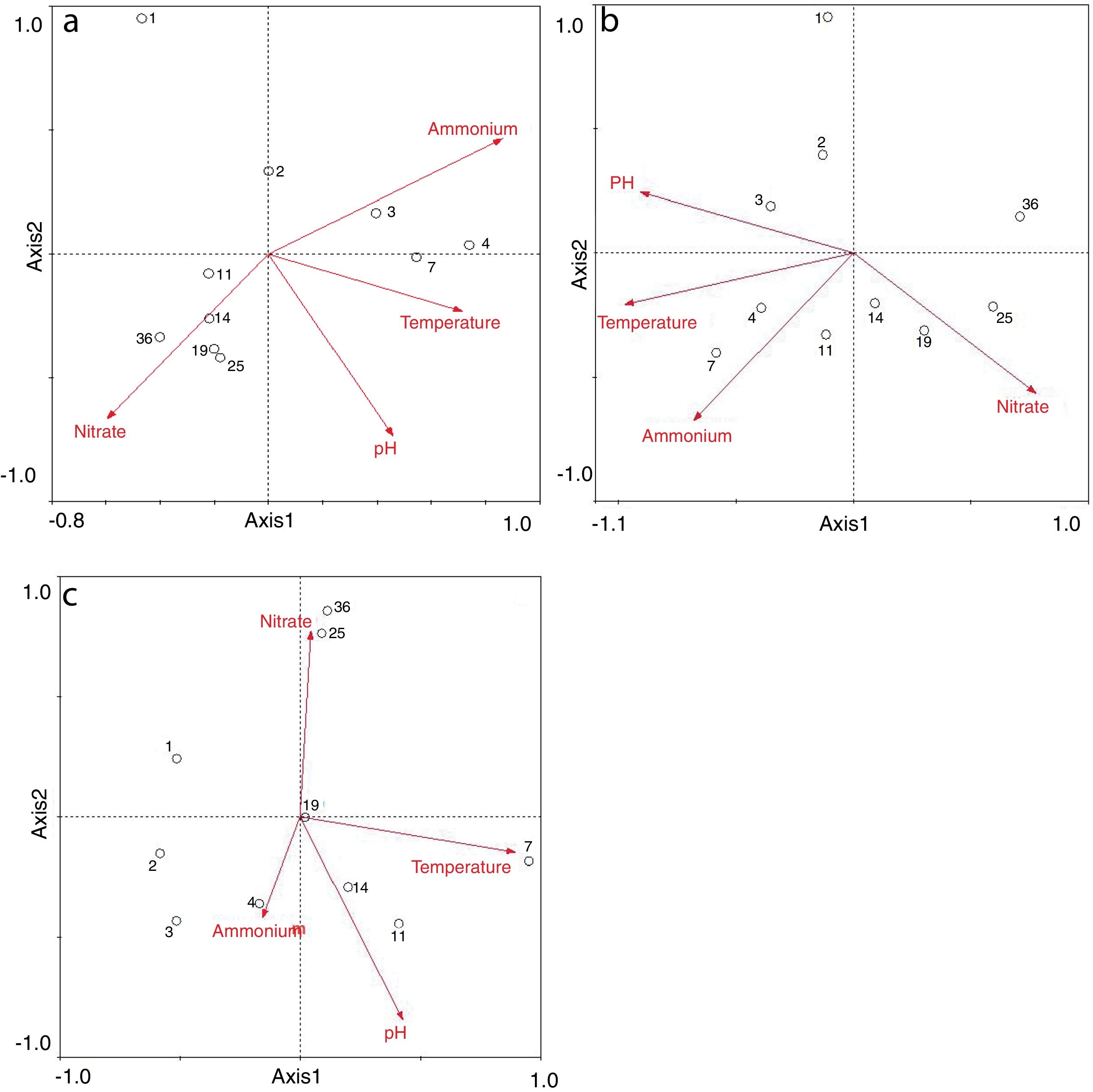
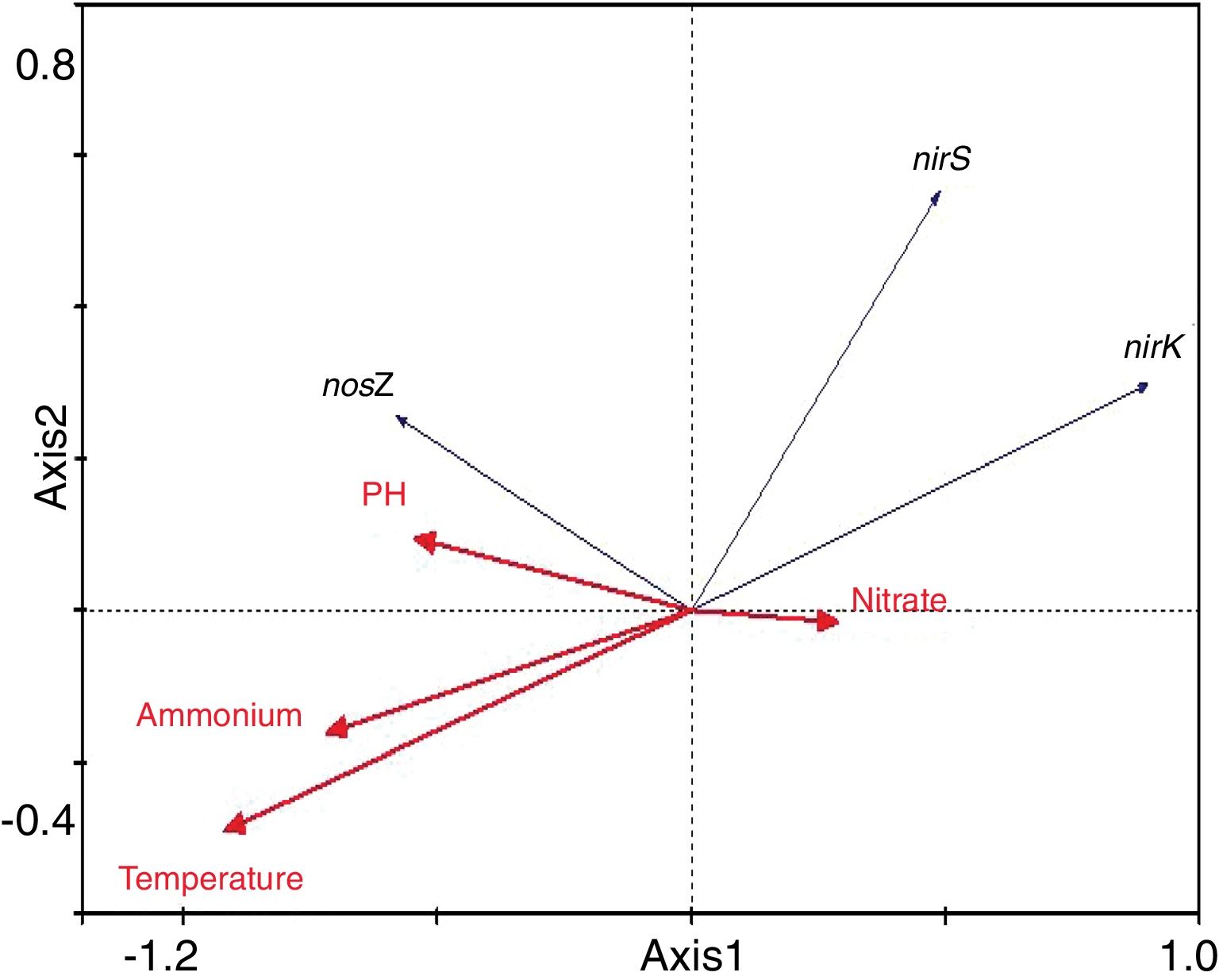
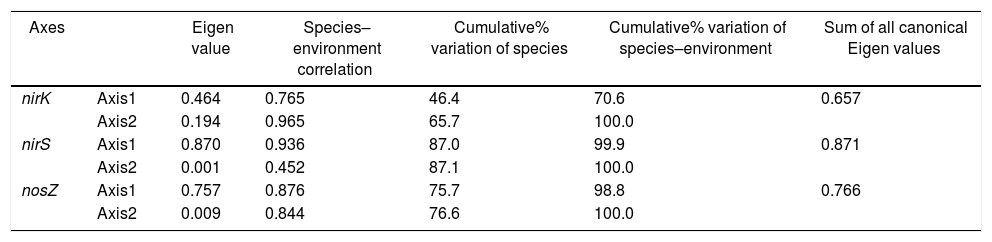
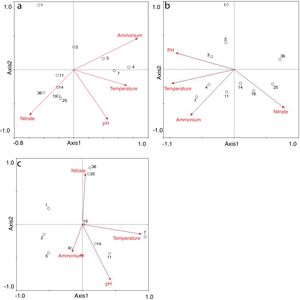
Redundancy analysis of diversity and abundanceIn order to investigate the impact of physico-chemical parameters (temperature, pH, NH4+–N and NO3−–N) on the denitrifier community in the compost, DGGE fingerprints of the nirK, nirS and nosZ genes were analyzed by redundancy analysis. The results are shown in Table 2. The first two canonical axes for the nirK gene explained 46.4% and 19.4% of the variation of community in the species data, respectively. For the nirS and nosZ genes, the first two canonical axes explained 87.0% and 75.7%, and 0.1% and 0.9% of the variation of community in the species data, respectively. Forward selection was performed to identify the parameters that best described the most influential gradients. Explanatory variables were added until the addition of further parameters failed to significantly improve the explanatory power of the model. In this procedure, NH4+–N was found to explain the significant variation (p<0.05) of the nirK and nirS gene species data. pH was found to explain the significant variation (p<0.05) of the nosZ gene species data. The results suggested that the variation of the three genes were affected by physico-chemical parameters. The positions of the samples collected with respect to the first two environmental axes are shown in Figure 5a for the nirK gene species data, in Figure 5b for the nirS gene species data, and in Figure 5c for the nosZ gene species data.
Redundancy analysis of DGGE profiles of the nirK, nirS and nosZ genes.
| Axes | Eigen value | Species–environment correlation | Cumulative% variation of species | Cumulative% variation of species–environment | Sum of all canonical Eigen values | |
|---|---|---|---|---|---|---|
| nirK | Axis1 | 0.464 | 0.765 | 46.4 | 70.6 | 0.657 |
| Axis2 | 0.194 | 0.965 | 65.7 | 100.0 | ||
| nirS | Axis1 | 0.870 | 0.936 | 87.0 | 99.9 | 0.871 |
| Axis2 | 0.001 | 0.452 | 87.1 | 100.0 | ||
| nosZ | Axis1 | 0.757 | 0.876 | 75.7 | 98.8 | 0.766 |
| Axis2 | 0.009 | 0.844 | 76.6 | 100.0 |
Monte Carlo permutation (n=499). Sum of all Eigen values for RDA was 1.000.
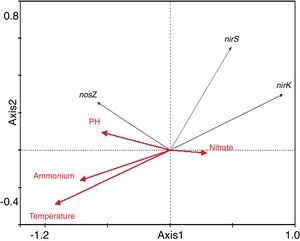
The results of the redundancy analysis for the abundance of the nirK, nirS and nosZ genes are shown in Table 3. The first two canonical axes explained 50.5% and 7.4% of the variation of abundance in the species data. Based on the results of the redundancy analysis, significant correlations (p<0.05) were found between the abundance of the nirK, nirS and nosZ genes and temperature (Fig. 6).
Redundancy analysis of abundance of the nirK, nirS and nosZ genes
| Axes | Eigen value | Species–environment correlation | Cumulative% variation of species | Cumulative% variation of species–environment | Sum of all canonical Eigen values |
|---|---|---|---|---|---|
| Axis1 | 0.505 | 0.880 | 50.5 | 87.3 | 0.579 |
| Axis2 | 0.074 | 0.461 | 57.9 | 100.0 |
Monte Carlo permutation (n=499). Sum of all Eigen values for RDA was 1.000.
The Shannon diversity index of the nirK, nirS and nosZ genes continuously increased during the first 7 days of composting (Table 1). This phenomenon could be explained by the fact that the relative abundance of organic compounds in the early stage of composting contribute to the growth of microorganisms and this may lead to a higher diversity of bacteria. In addition, accumulated ammonium and nitrate were converted into nitrite, which was necessary for the growth of denitrifiers. However, the highest abundance of denitrifying genes was detected on day 11 (Fig. 1). A similar phenomenon was also observed in pig manure composting33, which may be caused by the rapid accumulation of nitrate in the first 11 days and could provide enough substrate for the growth of denitrifiers.
It was observed that the diversity of the nirK gene was higher than that of the nirS gene during the whole composting process in this study. A similar phenomenon was also found in turfgrass soil11. Moreover, the abundance of the nirK gene was also higher than that of the nirS gene in this study, which suggested that nirK-type denitrifiers played a more important role in denitrification during composting than nirS-type denitrifiers. Similar results were obtained in a fertilized soil and a rice paddy field soil, where the nirK gene copy number was higher than that of the nirS gene14,24. It is interesting that in another report, Abell et al.1 found that nirS-type denitrifiers dominated the denitrifying process in estuary sediment. The copy number of the nirS gene in this study was higher than that in the sediments of a subtropical estuary4, suggesting that the composting environment may be more suitable for nirS-type denitrifiers than sediments. Abundance of the nosZ gene was higher than that of the nirS gene, but lower than that of the nirK gene in the compost in this study. However, no significant difference was found between the abundance of the nosZ gene and that of the nirK gene in the agricultural soil of France15, and the copy number of the nirS gene was higher than that of the nosZ gene in sediment soil8. Furthermore, the abundance of the nosZ gene was also lower than that in pasture soil2 and similar to that in agricultural field10. These findings suggest that different ecological conditions may favor different types of denitrifying genes (nirS gene and nirK gene).
According to the cloning and sequence analysis, denitrifiers obtained in this study were affiliated to Alphaproteobacteria (Rhodospirillales, Rhizobiales), Betaproteobacteria (Burkholderiales) and Gammaproteobacteria (Alteromonadales, Pseudomonadales), indicating that a wide variety of bacteria were involved in the process of denitrification during composting. This observation is also supported by the findings in other environments27,35. The result in this study could further confirm the conclusion that denitrifying bacteria are phylogenetically diverse in nature.
Correlation between physico-chemical parameters and denitrifying genesIn this study the redundancy analysis showed that the diversity of communities harboring nirK, nirS and nosZ genes was significantly correlated with ammonium and pH. In the presence of oxygen, ammonium can be oxidized to nitrate during the composting process, subsequently becoming substrate of denitrifiers under anaerobic conditions. Therefore, it is supposed that ammonium is crucial for the survival and growth of denitrifiers and has a significant impact on the diversity of the denitrifying community in the compost. However, no significant connection was found between the denitrifying community and ammonium in soil19, suggesting that the impacts of ammonium on denitrifying bacteria may vary under different ecological environments. pH plays a crucial role in the metabolism of microorganisms by affecting enzyme activity and the availability of nutrients. Therefore, pH has a prominent impact on the denitrifying community in different environment9. A significant correlation between pH and the nosZ community composition was also found during composting in the present study. This result is in accordance with Chen's5 report who found that pH variation was closely related to the denitrifying community in the composting process. In addition, under other ecological environments, such as in fen soil and grassland, pH also played an important role in the denitrification process9,26.
Temperature is crucial for growth of microorganisms. Not only population dynamics but also microbial metabolism is highly temperature-dependent21. Temperature also plays an important role in controlling the composting process because of its effect on the microbial population structure29. In the present study, the abundance of the nirK, nirS and nosZ genes was significantly correlated with temperature. However, pH, nitrate and ammonium, were not as significant as temperature in terms of influencing abundance of denitrifying gene abundance in this composting process. This finding is partly supported by the investigation conducted by Kandeler17, who found little correlation between abundance of denitrifiers and nitrate in the frozen soil layer.
In conclusion, diversity of nirK, nirS and nosZ genes reached the peak value on the thermophilic stage of cow manure composting and the diversity of the nirK gene is higher than that of nirS and nosZ genes. A higher abundance of nirK, nirS and nosZ genes was detected during the late thermophilic stage of composting and the abundance of the nirK gene was higher than that of nirS and nosZ genes in all the stages of composting. Ammonium and pH had a significant effect on diversity of nirK, nirS and nosZ genes, while temperature had a significant correlation with the abundance of the three genes.
FundingNational Natural Science Fund of China, project number 31372351 and 31672469.
Conflict of interestThe authors declare that they have no conflicts of interest.
This research project was supported by the National Natural Science Fund of China, project number 31372351 and 31672469. Thank all the authors for their contributions to the manuscript.