In Argentina, the epidemiological and molecular characteristics of Chlamydia psittaci infections are still not sufficiently known. A total of 846 respiratory and 10 ocular samples from patients with suspected human psittacosis were tested for C. psittaci from January 2010 to March 2015. Four samples of birds related to these patients were also studied. Forty-eight samples were positive for C. psittaci by a nested PCR. The molecular characterization of twelve C. psittaci PCR-positive samples received in the National Reference Laboratory INEI-ANLIS “Dr. Carlos G. Malbrán”, Buenos Aires, Argentina was performed. Eight positive samples from humans and four from birds were genotyped by ompA gene sequencing. C. psittaci genotype A was found in all human samples and in the related birds. This report contributes to our increasing knowledge of the epidemiological and molecular characteristics of C. psittaci to conduct effective surveillance of its zoonotic infections.
En la Argentina, aún no se conocen suficientemente las características epidemiológicas y moleculares de las infecciones por Chlamydia psittaci. Entre enero del 2010 y marzo del 2015 se estudiaron 846 muestras respiratorias y 10 oculares de pacientes con sospecha de psitacosis para la búsqueda de C. psittaci. También se estudiaron 4 muestras de aves relacionadas con estos pacientes. De ese total, 48 muestras fueron positivas para C. psittaci mediante una reacción en cadena de la polimerasa (PCR) anidada. Posteriormente, se realizó en el INEI-ANLIS «Dr. Carlos G. Malbrán» la caracterización molecular de 12 muestras positivas para C. psittaci, 8 de humanos y 4 de aves, que fueron genotipificadas por secuenciación del gen ompA. C. psittaci genotipo A se encontró en todas esas muestras. Este informe contribuye a mejorar nuestro conocimiento de las características epidemiológicas y moleculares de C. psittaci para lograr una vigilancia efectiva de la zoonosis que produce.
Psittacosis is a zoonosis caused by Chlamydia psittaci, an obligate intracellular bacterium belonging to the Chlamydiaceae family and its single genus, Chlamydia13.
Human infections vary from unapparent to severe systemic diseases: influenza – like illnesses, severe atypical pneumonia, endocarditis, myocarditis, meningitis, conjunctivitis, and others. The disease is rarely fatal in properly treated patients. Therefore, awareness of an early diagnosis is important. Pulmonary involvement is common and information of recent exposure to birds is frequently omitted during medical consultation.
Confirmation by molecular methods and genotyping of C. psittaci infection using human respiratory samples and animal samples is a matter of both diagnostic and epidemiological relevance6.
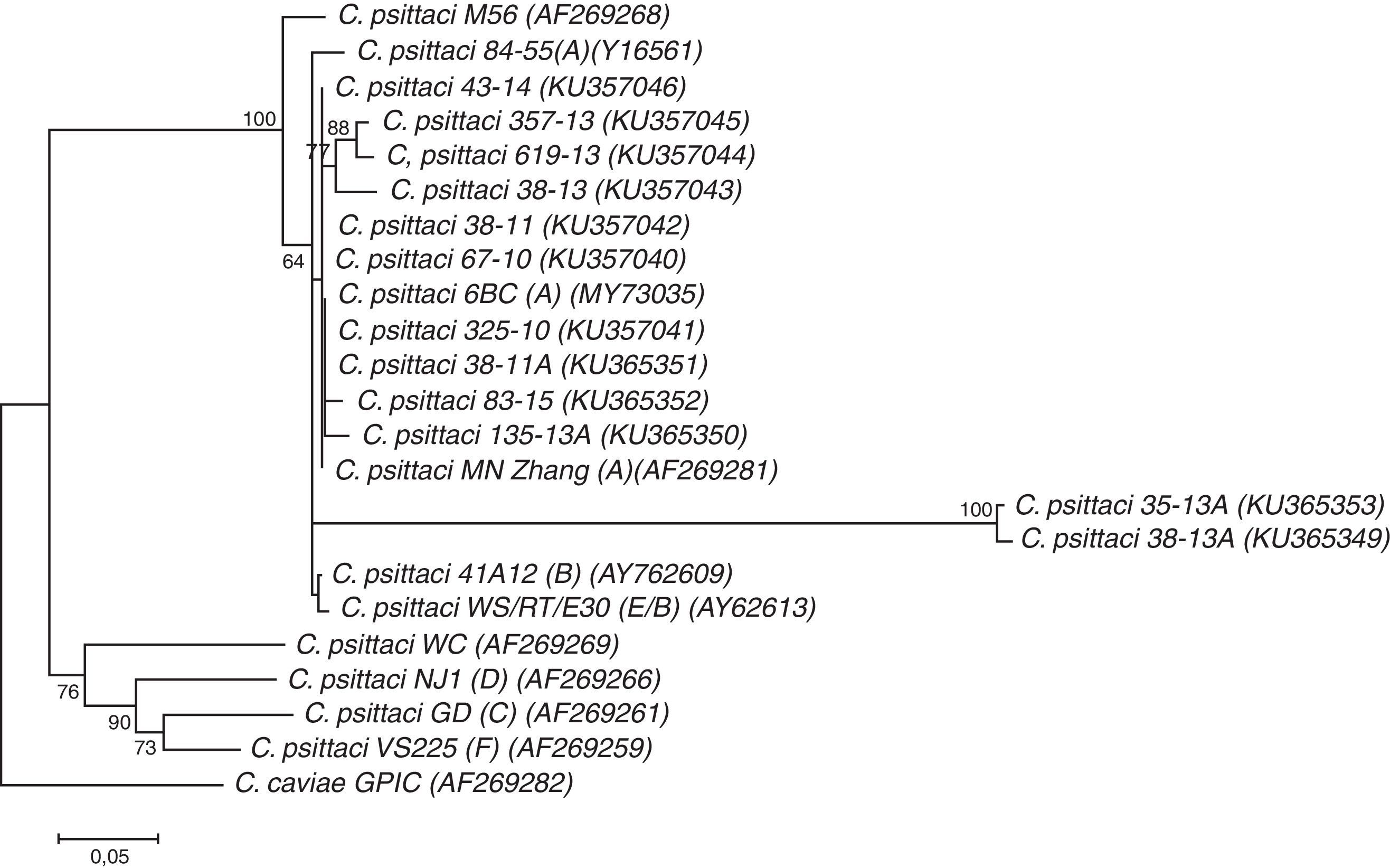
C. psittaci is divided into at least 9 genotypes (A–F, E/B, M56 and WC). Sequence analysis of the ompA gene which encodes the major outer membrane protein (MOMP) gene, is one way to identify all known and eventual new genotypes3.
Genotypes A–F and E/B are associated with birds and genotypes WC and M56 have been found in cattle and muskrats, respectively3. Genotype A is associated with Psittaciformes, B with pigeons, C with ducks and geese11, D with turkeys, E with pigeons and ratites, F with psittacine birds and turkeys5, and genotype E/B is mainly associated with ducks, turkeys and pigeons3,7.
The aim of this study was the molecular characterization of C. psittaci polymerase chain reaction-positive (PCR-positive) samples received in the National Reference Laboratory INEI-ANLIS “Dr. Carlos G. Malbrán”, Buenos Aires, Argentina.
Eight hundred and fifty-six samples from patients of different cities of Argentina, in whom psittacosis was suspected based on clinical presentation and/or history of exposure with infected birds, were analyzed between January 2010 and March 2015. Eight hundred and forty-six were respiratory samples (nasal and pharyngeal swabs, nasopharyngeal aspirates, tracheal aspirates and bronchioalveolar lavages) and 10 were ocular swabs. Cloacal and ocular swabs from 10 related birds were collected and stored until processed. Dacron swabs were placed in a 2ml sucrose–phosphate–glutamate or UTM (Copan Italia, Brescia, Italy), and stored at 4°C. Two hundred microliters of this suspension were subjected to DNA extraction using the DNeasy Blood & Tissue Kit (Qiagen, Valencia, CA, and USA) according to the manufacturer's instructions. DNA was eluted into 200μl of Qiagen elution buffer and stored at –20°C until tested.
A nested PCR protocol was used to amplify a portion of the 16S rRNA gene as previously described by Messmer et al.10 Briefly, 5μl of DNA extract was used in the first step to amplify a 436bp fragment common to the Chlamydia genus. In a second PCR step, 5μl of the PCR product obtained in the previous PCR reaction was used to amplify a 127bp specific product of C. psittaci.
Genotyping of C. psittaci was attempted in all positive samples in the 16S rRNA gene PCR. Amplification by nested PCR followed by sequencing of the variable domains III and IV of the ompA gene of C. psittaci was performed as described by Sachse and Hotzel.12 Briefly, 5μl of DNA extract were used in the first step to amplify a 576bp fragment of the ompA gene common to the Chlamydia genus. In a second PCR step, 5μl of the PCR product obtained in the previous PCR reaction were used to amplify a 389bp specific product of C. psittaci.
Nested PCR products were purified by gel electrophoresis using the AccuPrep® PCR Purification Kit (USA Bioneer Inc.) and subjected to a direct nucleotide sequencing reaction in both directions using the internal (second-round) PCR primers by Macrogen, Inc. (Seoul, Korea). The sequences obtained from fragments of the ompA gene were edited and prepared with BioEdit V 7.0.94 and subsequently aligned with Clustal X 2.128, along with the sequences downloaded from GenBank. Relatedness of newly characterized sequences was assessed by analysis using the 2.2.19 Basic Local Alignment Search Tool. The choice of the genomic region was based on the fact that it is widely associated with the genetic divergence of C. psittaci and it identifies all known genotypes3.
The dendrogram was constructed using the Tree Explorer module of the MEGA software version 414 with the neighbor-joining method and the p-distance parameter. The branch support was evaluated by nonparametric bootstrapping with 1000 pseudo-replicas.
Among the respiratory and ocular swabs samples of the 856 patients studied, 48 (5.6%) tested positive for C. psittaci by nested-PCR. Additionally, 6 samples from related birds tested positive.
Among these 54 positive samples for C. psittaci, only 12 produced enough PCR products in the ompA nested PCR to allow sequencing and genotype determination13 (8 from humans and 4 from birds). The mean age of the infected patients was 33.5 years (r=5–62 years); 62.5% (5/8) were young workers aged 23–45 years. Male/female ratio was 1:1.
The analysis of the medical records showed that community-acquired pneumonia (CAP) was the most frequent clinical presentation (50%; 4/8), followed by acute respiratory tract infection (25%; 2/8). Whooping cough was suspected in one case (12.5%; 1/8). One patient (12.5%; 1/8) with conjunctivitis tested positive for C. psittaci.
Five patients reported contact with birds, especially parrots (n=4). Contact with a domestic or companion bird was not reported in the remaining three C. psittaci positive patients.
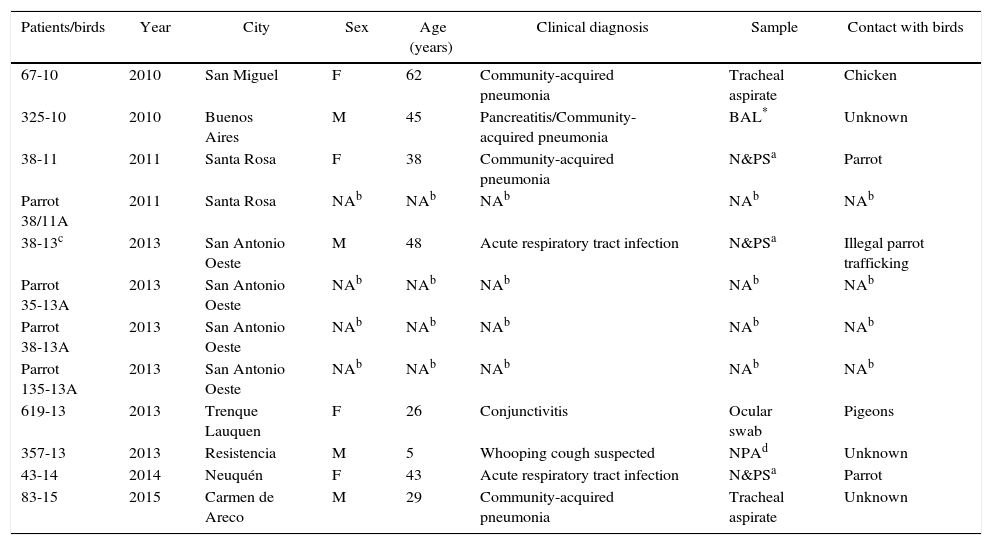
Clinical presentation and sources of the eight C. psittaci-positive samples are shown in Table 1.
Characteristics of the C. psittaci-positive cases
| Patients/birds | Year | City | Sex | Age (years) | Clinical diagnosis | Sample | Contact with birds |
|---|---|---|---|---|---|---|---|
| 67-10 | 2010 | San Miguel | F | 62 | Community-acquired pneumonia | Tracheal aspirate | Chicken |
| 325-10 | 2010 | Buenos Aires | M | 45 | Pancreatitis/Community-acquired pneumonia | BAL* | Unknown |
| 38-11 | 2011 | Santa Rosa | F | 38 | Community-acquired pneumonia | N&PSa | Parrot |
| Parrot 38/11A | 2011 | Santa Rosa | NAb | NAb | NAb | NAb | NAb |
| 38-13c | 2013 | San Antonio Oeste | M | 48 | Acute respiratory tract infection | N&PSa | Illegal parrot trafficking |
| Parrot 35-13A | 2013 | San Antonio Oeste | NAb | NAb | NAb | NAb | NAb |
| Parrot 38-13A | 2013 | San Antonio Oeste | NAb | NAb | NAb | NAb | NAb |
| Parrot 135-13A | 2013 | San Antonio Oeste | NAb | NAb | NAb | NAb | NAb |
| 619-13 | 2013 | Trenque Lauquen | F | 26 | Conjunctivitis | Ocular swab | Pigeons |
| 357-13 | 2013 | Resistencia | M | 5 | Whooping cough suspected | NPAd | Unknown |
| 43-14 | 2014 | Neuquén | F | 43 | Acute respiratory tract infection | N&PSa | Parrot |
| 83-15 | 2015 | Carmen de Areco | M | 29 | Community-acquired pneumonia | Tracheal aspirate | Unknown |
Genetic diversity and associations among the detected positive samples of C. psittaci were determined by the sequencing and genetic analysis of the composition of the ompA gene. The sequences obtained in this study were deposited in GenBank under accession numbers KU357040–KU357046; KU357049; KU365350–KU365353.
In the present study all C. psittaci-positive samples (from humans and birds) were recognized as belonging to genotype A by phylogenetic analysis (Fig. 1). However, in Córdoba, a central region of Argentina, genotype A was detected in only one case (12.5%, 1/8), being WC (75%, 6/8) the genotype most frequently found in patients with suspected human psittacosis1 and in captive birds2. Probably the differences could be due to different clinical conditions of the patients in both studies or/and to geographical sources. This differential genotypic profile is an interesting finding that should be investigated in the future. C. psittaci genotype A is more often found among psittacine birds such as parrots and cockatiels. However, the most prevalent C. psittaci genotype in human infections is currently unknown6.
Neighbor-joining dendrogram based on comparison of 290bp fragment of the ompA gene in Chlamydia. Numbers above branches are bootstrap values as a percentage of 1000 pseudo-replicates and only bootstrap values >60% are shown. Chlamydia caviae GPIC was used as an out-group. Scale bar shows the percentage sequence diversity.
The analysis of the medical records showed that CAP was the most frequent clinical presentation. In most studies on CAP, psittacosis was diagnosed only with serological tests. PCR leads to a quick diagnosis and lacks broad genus cross-reactivity. Furthermore, PCR products can be used to genotype C. psittaci, which is relevant for public health notification, source detection and control.
The use of nucleic acid amplification directly from clinical samples and direct genotyping with phylogenetic analysis in the ompA gene could improve the surveillance and molecular epidemiology of this disease. In this sense, the classical method applying two PCR techniques plus sequencing was successfully used and it should be implemented in regional laboratories for an improved diagnosis of this infectious disease.
In this study, a nested PCR and direct sequencing were used to detect and genetically characterize C. psittaci. For the phylogenetic analysis of C. psittaci, we used partial ompA gene amplification. Our results only report the circulation of genotype A associated with respiratory symptoms and conjunctivitis in different areas of our country, at least in our experience with these eight human cases. Psittaciformes especially parakeets and parrots, and individuals in contact with them or with racing pigeons, were the most frequently infected.
C. psittaci is rarely suspected in ocular infections, but using molecular techniques, as we did, may be more commonly found9. In the present study we showed a case of conjunctivitis due to C. psittaci in a 26 year-old woman not responding to the current treatment and living in contact with wild birds.
Several limitations of our study need to be considered. Isolation in cell cultures was the preferred technique to confirm and corroborate the results obtained; however, in the case of C. psittaci, this procedure is unadvisable because of the biological risks. Moreover, the phylogenetic analysis showed two sequences corresponding to birds (KU365353 and KU365349) in separated clusters. As can be seen in Figure 1 they have a low degree of homology with others obtained in the same city (74% similarity, one with an avian strain and the other with a human strain) (data not shown). Further studies are needed to confirm the genotype and even the species.
Effective public health surveillance for new variants and modes of transmission will be helpful to understanding the evolution and epidemiology of these bacteria. Therefore, new information about sequence variation of subtypes will be needed.
Ethical responsibilitiesProtection of human and animal subjectsThe authors state that for this investigation no experiments have been performed on humans or animals.
Confidentiality of dataThe authors state that they have followed the protocols of their work center on the publication of patient data.
Right to privacy and informed consentThe authors state that in this article there are no patient data.
Conflicts of interestThe authors declare that they have no conflicts of interest.
This study was funded by PICTO-ANLIS 2011-0180, Agencia Nacional de Promoción Científica y Tecnológica del Ministerio de Ciencia, Tecnología e Innovación Productiva de la República Argentina. MSc María Estela Cadario is a staff member of the Instituto Nacional de Enfermedades Infecciosas-ANLIS “Dr Carlos G Malbrán”, Buenos Aires, Argentina.