This study aimed to determine the clonal relationship among 137 Streptococcus uberis isolates from bovine milk with subclinical or clinical mastitis in Argentina and to assess the prevalence and conservation of pauA and sua genes. This information is critical for the rational design of a vaccine for the prevention of bovine mastitis caused by S. uberis. The isolates were typed by random amplified polymorphic DNA (RAPD) analysis and by pulsed-field gel electrophoresis (PFGE). The 137 isolates exhibited 61 different PFGE types and 25 distinct RAPD profiles. Simpson's diversity index was calculated both for PFGE (0.983) and for RAPD (0.941), showing a high discriminatory power in both techniques. The analysis of the relationship between pairs of isolates showed 92.6% concordance between both techniques indicating that any given pair of isolates distinguished by one method tended to be distinguished by the other. The prevalence of the sua and pauA genes was 97.8% (134/137) and 94.9% (130/137), respectively. Nucleotide and amino acid sequences of the sua and pauA genes from 20 S. uberis selected isolates, based on their PFGE and RAPD types and geographical origin, showed an identity between 95% and 100% with respect to all reference sequences registered in GenBank. These results demonstrate that, in spite of S. uberis clonal diversity, the sua and pauA genes are prevalent and highly conserved, showing their importance to be included in future vaccine studies to prevent S. uberis bovine mastitis.
Este estudio pretendió determinar la relación clonal entre 137 aislamientos de S. uberis obtenidos de leche de bovinos con mastitis clínica o subclínica en la Argentina, como así también la prevalencia y la conservación de los genes sua y PauA entre dichos aislamientos. Esta información es crítica para el diseño racional de una vacuna que prevenga la mastitis bovina por S. uberis. Los aislamientos se tipificaron molecularmente por amplificación al azar del ADN polimórfico (RAPD) y mediante electroforesis de campos pulsados (PFGE). Los 137 aislamientos mostraron 61 pulsotipos mediante PFGE y 25 tipos de RAPD diferentes. Los índices de Simpson calculados fueron 0,983 por PFGE y 0,941 por RAPD; esto evidencia el elevado poder discriminatorio de ambas técnicas. El análisis de la relación entre pares de aislamientos mostró un 92,6% de concordancia entre ambas técnicas, lo que indica que cualquier par de aislamientos que fue distinguido por un método tendió a ser distinguido por el otro. La prevalencia de los genes sua y puaA fue del 97,8% (134/137) y 94,9% (130/137), respectivamente. Las secuencias de nucleótidos y de aminoácidos codificados por los genes sua y pauA de los 20 aislamientos de S. uberis seleccionados sobre la base de su tipo de PFGE y RAPD y origen geográfico tuvieron un porcentaje de identidad de entre 95% y 100% con respecto a todas las secuencias de referencia registradas en GenBank. Estos resultados demuestran que, a pesar de la diversidad clonal de S. uberis, los genes sua y pauA son prevalentes y están altamente conservados y deberían ser incluidos en futuros estudios de vacunas para prevenir mastitis bovina causada por S. uberis.
Bovine mastitis is one of the most costly diseases of dairy cattle as a consequence of antibiotic treatment expenses, decreased milk production and quality, and increased animal replacement rate6,30. In Argentina, milk yield losses in cows suffering from mastitis were reported to be of about $4.3/cow/days41. Bovine intramammary infections (IMI) are caused by both contagious and environmental bacteria. Streptococcus uberis is one of the most prevalent environmental pathogens associated with subclinical and clinical IMI both in lactating and non-lactating cows7,27. In addition, this pathogen can persist in the mammary gland causing chronic IMI47. Traditional control procedures based on milking time hygiene and antibiotic therapy are considered adequate to reduce incidence of most contagious pathogens, but are often insufficient for the control of IMI caused by S. uberis20. Consequently, the interest has focused on the development of immunoprophylactic strategies. However, a significant obstacle in the design of an effective vaccine is the high level of genetic variability of different isolates of S. uberis, frequently involving virulence factor genes18,31. In this context, epidemiological studies of regional isolates are extremely useful to detect the frequency and distribution of bacterial types associated with IMI and to identify target molecules for the development of immunogens and therapeutic agents. Methods based on DNA analysis including restriction fragment length polymorphism (RFLP), random amplified polymorphic DNA (RAPD), pulsed field gel electrophoresis (PFGE), and multilocus sequence typing (MLST) have been successfully applied to fulfill this need. PFGE is the most discriminatory method for typing bacteria22, whereas RAPD is a more straightforward method given its relatively lower costs, execution time demanded, less expensive equipment requirements and sensitivity. Both methods have been widely used to study the genetic variability of many bacterial species, including important human pathogens22,37.
Several S. uberis virulence factors have been described16,20,23,24. Among them, plasminogen activator A (PauA)35 and S. uberis adhesion molecule (SUAM)2 have shown potential as protective immunogens. The former is a protease capable of activating plasmin, which in turn, degrades proteins producing small peptides and free amino acids used by bacteria as a nitrogen source. This factor has been related to early mammary gland colonization21 while SUAM both to adherence and internalization, through its binding to lactoferrin28, and to bacterial persistence in bovine mammary epithelial cells in vitro2.
The wide genetic diversity observed in S. uberis indicates that virulence is not associated with any specific molecular type43. Therefore, immunologic prevention strategies against this organism should be directed to a multitude of different factors included in field isolates from individual herds. Nevertheless, the difficulty associated with S. uberis genetic variability can be currently overcome through bioinformatic tools that allow to analyze DNA sequence encoding genes from virulence factors which are found in a large number of isolates. Thus, epidemiological studies on gene distribution together with their sequence analysis will contribute to identify potential antigens as vaccine components.
The main objectives of this work were to determine the clonal relationship between S. uberis isolates from milk of cows suffering from subclinical or clinical mastitis in Argentina and to assess the prevalence and conservation of the pauA and sua genes.
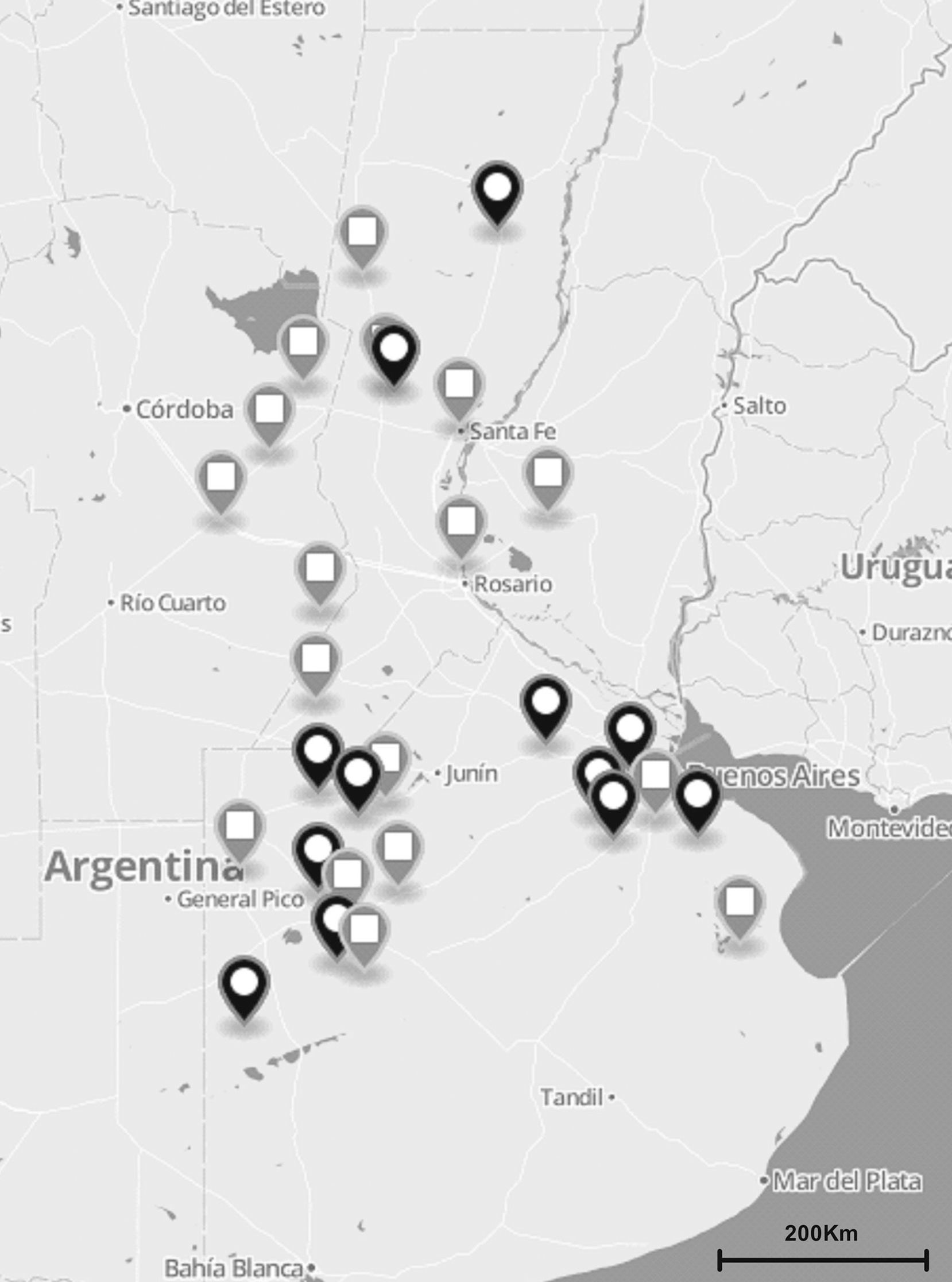
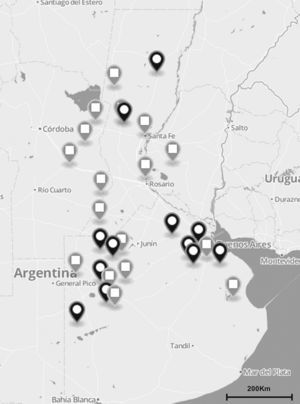
Materials and methodsS. uberis collection and DNA preparationA total of 137 S. uberis strains were isolated from milk of 122 cows suffering from clinical and subclinical mastitis between November 2010 and July 2012. Milk samples were obtained from 35 dairy herds located in the main dairy regions of Argentina (Fig. 1). Clinical mastitis was characterized by evidence of inflammation (abnormal milk, heat, swelling or pain of affected quarters) while subclinical mastitis by lack of local or systemic signs of mammary inflammation and somatic cell counts ≥200,000cells/ml.
The total number (n=137) of S. uberis isolates recovered from bovines suffering from subclinical or clinical mastitis were from 35 dairy farms located in the main dairy region of Argentina (black and gray globes). Gray globes indicate geographical origin of isolates selected for the sua and pauA sequencing.
S. uberis isolates were initially identified by standard conventional biochemical tests such as esculin hydrolysis, hippurate hydrolysis, growth on 6.5% NaCl, growth on bile-esculin-agar and the CAMP test25. Restriction fragment length polymorphism (RFLP) of 16S rDNA14 was performed for molecular confirmation. S. uberis ATCC 27958 was used as reference strain.
Genomic DNA was extracted using a modification of the method described by Hill and Leigh13. The detailed protocol is available at pubmlst.org/suberis/info/protocol.shtml.
Molecular typing of S. uberis strainsMolecular typing was carried out by PFGE and RAPD-PCR. Pulsed-field electrophoresis was performed in accordance with a previously described method5. Briefly, electrophoresis was performed with the CHEF-DR III SYSTEM (Bio-Rad Laboratories) using 1% Pulsed Field Certified™ (BioRad Laboratories) in 0.5× Tris-borate-EDTA (TBE) buffer at 11.3°C for 23h. The electrophoresis conditions were as follows: initial switch time value of 5s, final switch time of 35s at a gradient of 6V/cm. After electrophoresis, the gel was stained with ethidium bromide solution (0.5mg/l) and then destained with deionized water. Then the DNA bands were visualized with a UV transilluminator and a digital image of PFGE patterns was obtained using Molecular Imager® Gel Doc™ XR+ System (BioRad, Laboratories Inc., Richmond, CA, USA).
RAPD-PCR was performed as previously described45 using primer OPE-4. The amplified products were separated by gel electrophoresis in 1.5% agarose gel, stained with ethidium bromide. Banding pattern similarities were analyzed using the Dice correlation coefficient. Values for tolerance and optimization were set at 1.5% and 1%, respectively. The cluster analysis was based on UPGMA, Unweighted Pair Group Method with Arithmetic averages, and data were analyzed with the TREECON software for Windows39. Isolates indicating more than 3 DNA fragment differences and a similarity of < 80% following the dendrogram analysis were classified as different PFGE or RAPD types, whereas fragment variations and a similarity of > 80% following the dendrogram analysis were defined as PFGE or RAPD subtypes. Isolates showing identical patterns (100% pattern similarity index) or a similarity index of > 80% were interpreted as belonging to the same PFGE or RAPD type1. The typing experiments were repeated at least twice.
Calculation of concordance, Simpson's index of diversity and Wallace's coefficientsSimpson's index of diversity was calculated to measure the discriminatory power of the typing systems. This index indicates the probability that 2 strains randomly sampled from a population will belong to 2 different types4. Wallace's coefficients were used to explore the correlation between results produced by PFGE and RAPD. These were calculated using EpiCompare version 1.0 (http://www3.ridom.de/epicompare/). Group concordance was evaluated by cross-classification of all possible pairs of isolates as previously described34. All possible pairs of isolates were cross-classified according to matched or mismatched types. The resulting 2×2 table was evaluated by the chi-square test and the percentage of concordant cells was calculated using the GraphPad Prism 5 (Version 5, USA) software.
PCRs and DNA sequencingDNA amplification for the sua gene was performed using oligonucleotide primers derived from published sequences23. Amplification of the pauA gene was performed using the following primers, which were designed using DNAstar Primer Select software (DNAstar, Madison, WI), 5′-TTTTTAATATTAATGCTTTTG-3′ and 5′-AGAAAAATTTAATGGATAC-3′. The reaction mixture was made with 150ng/μl template DNA, 0.8μM oligonucleotide primers, 0.2μM of each of the four dNTPs, 1.25U Taq polymerase and 3mM MgCl2, in a final volume of 50μl. The expected amplicon size was 874bp. PCR conditions were 5min at 95°C, 30 cycles of [1min at 94°C, 1min at 49°C and 1min at 72°C], 5min at 72°C.
The PCR products from 20 isolates of S. uberis were selected for sequencing. The following criteria were taken into account for isolate selection: isolates had to belong to different geographical regions and/or had to be epidemiologically unrelated (Table 1; Figs. 1 and 2). DNA was purified using the Wizard®SV Gel and PCR Clean-Up System (Promega) according to the manufacturers’ protocol and eluted in 30μl of sterile MilliQ water. Positive PCR products were sequenced in both directions using the same specific PCR primers for the pauA gene while for the sua gene the following oligonucleotides were added: sua2: 5′-GAA TTC ACA CAA TCT GAC GAG GT-3′; sua3: 5′-GAA TTC GAA GTT GGG GCA TAC-3′ and sua4: 5′-GAA TTC CCA AGT GCT CCG GTC T-3′ to the PCR primers. DNA samples (30–50ng/μl) were sequenced by ABI3130XL sequencer analyzer (Applied Biosystems) from the sequencing service of the Biotechnology Institute of INTA Castelar, Argentina.
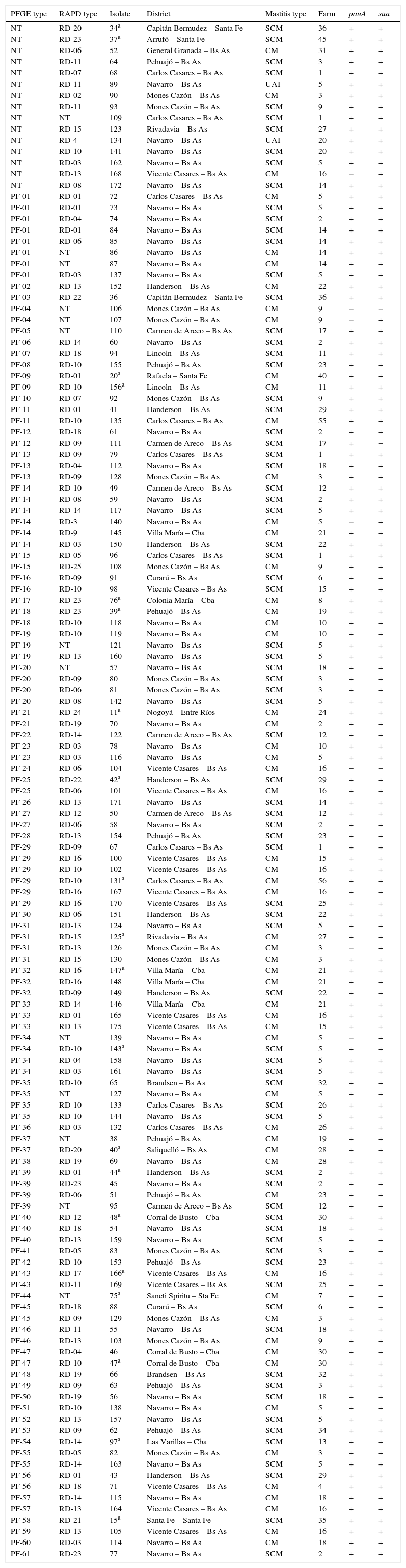
Characteristics of S. uberis isolates according to their PFGE type.
| PFGE type | RAPD type | Isolate | District | Mastitis type | Farm | pauA | sua |
|---|---|---|---|---|---|---|---|
| NT | RD-20 | 34a | Capitán Bermudez – Santa Fe | SCM | 36 | + | + |
| NT | RD-23 | 37a | Arrufó – Santa Fe | SCM | 45 | + | + |
| NT | RD-06 | 52 | General Granada – Bs As | CM | 31 | + | + |
| NT | RD-11 | 64 | Pehuajó – Bs As | SCM | 3 | + | + |
| NT | RD-07 | 68 | Carlos Casares – Bs As | SCM | 1 | + | + |
| NT | RD-11 | 89 | Navarro – Bs As | UAI | 5 | + | + |
| NT | RD-02 | 90 | Mones Cazón – Bs As | CM | 3 | + | + |
| NT | RD-11 | 93 | Mones Cazón – Bs As | SCM | 9 | + | + |
| NT | NT | 109 | Carlos Casares – Bs As | SCM | 1 | + | + |
| NT | RD-15 | 123 | Rivadavia – Bs As | SCM | 27 | + | + |
| NT | RD-4 | 134 | Navarro – Bs As | UAI | 20 | + | + |
| NT | RD-10 | 141 | Navarro – Bs As | SCM | 20 | + | + |
| NT | RD-03 | 162 | Navarro – Bs As | SCM | 5 | + | + |
| NT | RD-13 | 168 | Vicente Casares – Bs As | CM | 16 | − | + |
| NT | RD-08 | 172 | Navarro – Bs As | SCM | 14 | + | + |
| PF-01 | RD-01 | 72 | Carlos Casares – Bs As | CM | 5 | + | + |
| PF-01 | RD-01 | 73 | Navarro – Bs As | SCM | 5 | + | + |
| PF-01 | RD-04 | 74 | Navarro – Bs As | SCM | 2 | + | + |
| PF-01 | RD-01 | 84 | Navarro – Bs As | SCM | 14 | + | + |
| PF-01 | RD-06 | 85 | Navarro – Bs As | SCM | 14 | + | + |
| PF-01 | NT | 86 | Navarro – Bs As | CM | 14 | + | + |
| PF-01 | NT | 87 | Navarro – Bs As | CM | 14 | + | + |
| PF-01 | RD-03 | 137 | Navarro – Bs As | SCM | 5 | + | + |
| PF-02 | RD-13 | 152 | Handerson – Bs As | CM | 22 | + | + |
| PF-03 | RD-22 | 36 | Capitán Bermudez – Santa Fe | SCM | 36 | + | + |
| PF-04 | NT | 106 | Mones Cazón – Bs As | CM | 9 | − | − |
| PF-04 | NT | 107 | Mones Cazón – Bs As | CM | 9 | − | + |
| PF-05 | NT | 110 | Carmen de Areco – Bs As | SCM | 17 | + | + |
| PF-06 | RD-14 | 60 | Navarro – Bs As | SCM | 2 | + | + |
| PF-07 | RD-18 | 94 | Lincoln – Bs As | SCM | 11 | + | + |
| PF-08 | RD-10 | 155 | Pehuajó – Bs As | SCM | 23 | + | + |
| PF-09 | RD-01 | 20a | Rafaela – Santa Fe | CM | 40 | + | + |
| PF-09 | RD-10 | 156a | Lincoln – Bs As | CM | 11 | + | + |
| PF-10 | RD-07 | 92 | Mones Cazón – Bs As | SCM | 9 | + | + |
| PF-11 | RD-01 | 41 | Handerson – Bs As | SCM | 29 | + | + |
| PF-11 | RD-10 | 135 | Carlos Casares – Bs As | CM | 55 | + | + |
| PF-12 | RD-18 | 61 | Navarro – Bs As | SCM | 2 | + | + |
| PF-12 | RD-09 | 111 | Carmen de Areco – Bs As | SCM | 17 | + | − |
| PF-13 | RD-09 | 79 | Carlos Casares – Bs As | SCM | 1 | + | + |
| PF-13 | RD-04 | 112 | Navarro – Bs As | SCM | 18 | + | + |
| PF-13 | RD-09 | 128 | Mones Cazón – Bs As | CM | 3 | + | + |
| PF-14 | RD-10 | 49 | Carmen de Areco – Bs As | SCM | 12 | + | + |
| PF-14 | RD-08 | 59 | Navarro – Bs As | SCM | 2 | + | + |
| PF-14 | RD-14 | 117 | Navarro – Bs As | SCM | 5 | + | + |
| PF-14 | RD-3 | 140 | Navarro – Bs As | CM | 5 | − | + |
| PF-14 | RD-9 | 145 | Villa María – Cba | CM | 21 | + | + |
| PF-14 | RD-03 | 150 | Handerson – Bs As | SCM | 22 | + | + |
| PF-15 | RD-05 | 96 | Carlos Casares – Bs As | SCM | 1 | + | + |
| PF-15 | RD-25 | 108 | Mones Cazón – Bs As | CM | 9 | + | + |
| PF-16 | RD-09 | 91 | Curarú – Bs As | SCM | 6 | + | + |
| PF-16 | RD-10 | 98 | Vicente Casares – Bs As | SCM | 15 | + | + |
| PF-17 | RD-23 | 76a | Colonia María – Cba | CM | 8 | + | + |
| PF-18 | RD-23 | 39a | Pehuajó – Bs As | CM | 19 | + | + |
| PF-18 | RD-10 | 118 | Navarro – Bs As | CM | 10 | + | + |
| PF-19 | RD-10 | 119 | Navarro – Bs As | CM | 10 | + | + |
| PF-19 | NT | 121 | Navarro – Bs As | SCM | 5 | + | + |
| PF-19 | RD-13 | 160 | Navarro – Bs As | SCM | 5 | + | + |
| PF-20 | NT | 57 | Navarro – Bs As | SCM | 18 | + | + |
| PF-20 | RD-09 | 80 | Mones Cazón – Bs As | SCM | 3 | + | + |
| PF-20 | RD-06 | 81 | Mones Cazón – Bs As | SCM | 3 | + | + |
| PF-20 | RD-08 | 142 | Navarro – Bs As | SCM | 5 | + | + |
| PF-21 | RD-24 | 11a | Nogoyá – Entre Ríos | CM | 24 | + | + |
| PF-21 | RD-19 | 70 | Navarro – Bs As | CM | 2 | + | + |
| PF-22 | RD-14 | 122 | Carmen de Areco – Bs As | SCM | 12 | + | + |
| PF-23 | RD-03 | 78 | Navarro – Bs As | CM | 10 | + | + |
| PF-23 | RD-03 | 116 | Navarro – Bs As | CM | 5 | + | + |
| PF-24 | RD-06 | 104 | Vicente Casares – Bs As | CM | 16 | − | − |
| PF-25 | RD-22 | 42a | Handerson – Bs As | SCM | 29 | + | + |
| PF-25 | RD-06 | 101 | Vicente Casares – Bs As | CM | 16 | + | + |
| PF-26 | RD-13 | 171 | Navarro – Bs As | SCM | 14 | + | + |
| PF-27 | RD-12 | 50 | Carmen de Areco – Bs As | SCM | 12 | + | + |
| PF-27 | RD-06 | 58 | Navarro – Bs As | SCM | 2 | + | + |
| PF-28 | RD-13 | 154 | Pehuajó – Bs As | SCM | 23 | + | + |
| PF-29 | RD-09 | 67 | Carlos Casares – Bs As | SCM | 1 | + | + |
| PF-29 | RD-16 | 100 | Vicente Casares – Bs As | CM | 15 | + | + |
| PF-29 | RD-10 | 102 | Vicente Casares – Bs As | CM | 16 | + | + |
| PF-29 | RD-10 | 131a | Carlos Casares – Bs As | CM | 56 | + | + |
| PF-29 | RD-16 | 167 | Vicente Casares – Bs As | CM | 16 | + | + |
| PF-29 | RD-16 | 170 | Vicente Casares – Bs As | SCM | 25 | + | + |
| PF-30 | RD-06 | 151 | Handerson – Bs As | SCM | 22 | + | + |
| PF-31 | RD-13 | 124 | Navarro – Bs As | SCM | 5 | + | + |
| PF-31 | RD-15 | 125a | Rivadavia – Bs As | CM | 27 | + | + |
| PF-31 | RD-13 | 126 | Mones Cazón – Bs As | CM | 3 | − | + |
| PF-31 | RD-15 | 130 | Mones Cazón – Bs As | CM | 3 | + | + |
| PF-32 | RD-16 | 147a | Villa María – Cba | CM | 21 | + | + |
| PF-32 | RD-16 | 148 | Villa María – Cba | CM | 21 | + | + |
| PF-32 | RD-09 | 149 | Handerson – Bs As | SCM | 22 | + | + |
| PF-33 | RD-14 | 146 | Villa María – Cba | CM | 21 | + | + |
| PF-33 | RD-01 | 165 | Vicente Casares – Bs As | CM | 16 | + | + |
| PF-33 | RD-13 | 175 | Vicente Casares – Bs As | CM | 15 | + | + |
| PF-34 | NT | 139 | Navarro – Bs As | CM | 5 | − | + |
| PF-34 | RD-10 | 143a | Navarro – Bs As | SCM | 5 | + | + |
| PF-34 | RD-04 | 158 | Navarro – Bs As | SCM | 5 | + | + |
| PF-34 | RD-03 | 161 | Navarro – Bs As | SCM | 5 | + | + |
| PF-35 | RD-10 | 65 | Brandsen – Bs As | SCM | 32 | + | + |
| PF-35 | NT | 127 | Navarro – Bs As | CM | 5 | + | + |
| PF-35 | RD-10 | 133 | Carlos Casares – Bs As | SCM | 26 | + | + |
| PF-35 | RD-10 | 144 | Navarro – Bs As | SCM | 5 | + | + |
| PF-36 | RD-03 | 132 | Carlos Casares – Bs As | CM | 26 | + | + |
| PF-37 | NT | 38 | Pehuajó – Bs As | CM | 19 | + | + |
| PF-37 | RD-20 | 40a | Saliquelló – Bs As | CM | 28 | + | + |
| PF-38 | RD-19 | 69 | Navarro – Bs As | CM | 28 | + | + |
| PF-39 | RD-01 | 44a | Handerson – Bs As | SCM | 2 | + | + |
| PF-39 | RD-23 | 45 | Navarro – Bs As | SCM | 2 | + | + |
| PF-39 | RD-06 | 51 | Pehuajó – Bs As | CM | 23 | + | + |
| PF-39 | NT | 95 | Carmen de Areco – Bs As | SCM | 12 | + | + |
| PF-40 | RD-12 | 48a | Corral de Busto – Cba | SCM | 30 | + | + |
| PF-40 | RD-18 | 54 | Navarro – Bs As | SCM | 18 | + | + |
| PF-40 | RD-13 | 159 | Navarro – Bs As | SCM | 5 | + | + |
| PF-41 | RD-05 | 83 | Mones Cazón – Bs As | SCM | 3 | + | + |
| PF-42 | RD-10 | 153 | Pehuajó – Bs As | SCM | 23 | + | + |
| PF-43 | RD-17 | 166a | Vicente Casares – Bs As | CM | 16 | + | + |
| PF-43 | RD-11 | 169 | Vicente Casares – Bs As | SCM | 25 | + | + |
| PF-44 | NT | 75a | Sancti Spiritu – Sta Fe | CM | 7 | + | + |
| PF-45 | RD-18 | 88 | Curarú – Bs As | SCM | 6 | + | + |
| PF-45 | RD-09 | 129 | Mones Cazón – Bs As | CM | 3 | + | + |
| PF-46 | RD-11 | 55 | Navarro – Bs As | SCM | 18 | + | + |
| PF-46 | RD-13 | 103 | Mones Cazón – Bs As | CM | 9 | + | + |
| PF-47 | RD-04 | 46 | Corral de Busto – Cba | CM | 30 | + | + |
| PF-47 | RD-10 | 47a | Corral de Busto – Cba | CM | 30 | + | + |
| PF-48 | RD-19 | 66 | Brandsen – Bs As | SCM | 32 | + | + |
| PF-49 | RD-09 | 63 | Pehuajó – Bs As | SCM | 3 | + | + |
| PF-50 | RD-19 | 56 | Navarro – Bs As | SCM | 18 | + | + |
| PF-51 | RD-10 | 138 | Navarro – Bs As | CM | 5 | + | + |
| PF-52 | RD-13 | 157 | Navarro – Bs As | SCM | 5 | + | + |
| PF-53 | RD-09 | 62 | Pehuajó – Bs As | SCM | 34 | + | + |
| PF-54 | RD-14 | 97a | Las Varillas – Cba | SCM | 13 | + | + |
| PF-55 | RD-05 | 82 | Mones Cazón – Bs As | CM | 3 | + | + |
| PF-55 | RD-14 | 163 | Navarro – Bs As | SCM | 5 | + | + |
| PF-56 | RD-01 | 43 | Handerson – Bs As | SCM | 29 | + | + |
| PF-56 | RD-18 | 71 | Vicente Casares – Bs As | CM | 4 | + | + |
| PF-57 | RD-14 | 115 | Navarro – Bs As | CM | 18 | + | + |
| PF-57 | RD-13 | 164 | Vicente Casares – Bs As | CM | 16 | + | + |
| PF-58 | RD-21 | 15a | Santa Fe – Santa Fe | SCM | 35 | + | + |
| PF-59 | RD-13 | 105 | Vicente Casares – Bs As | CM | 16 | + | + |
| PF-60 | RD-03 | 114 | Navarro – Bs As | CM | 18 | + | + |
| PF-61 | RD-23 | 77 | Navarro – Bs As | SCM | 2 | + | + |
CM, clinical mastitis; SCM, subclinical mastitis; UAI, unavailable information.
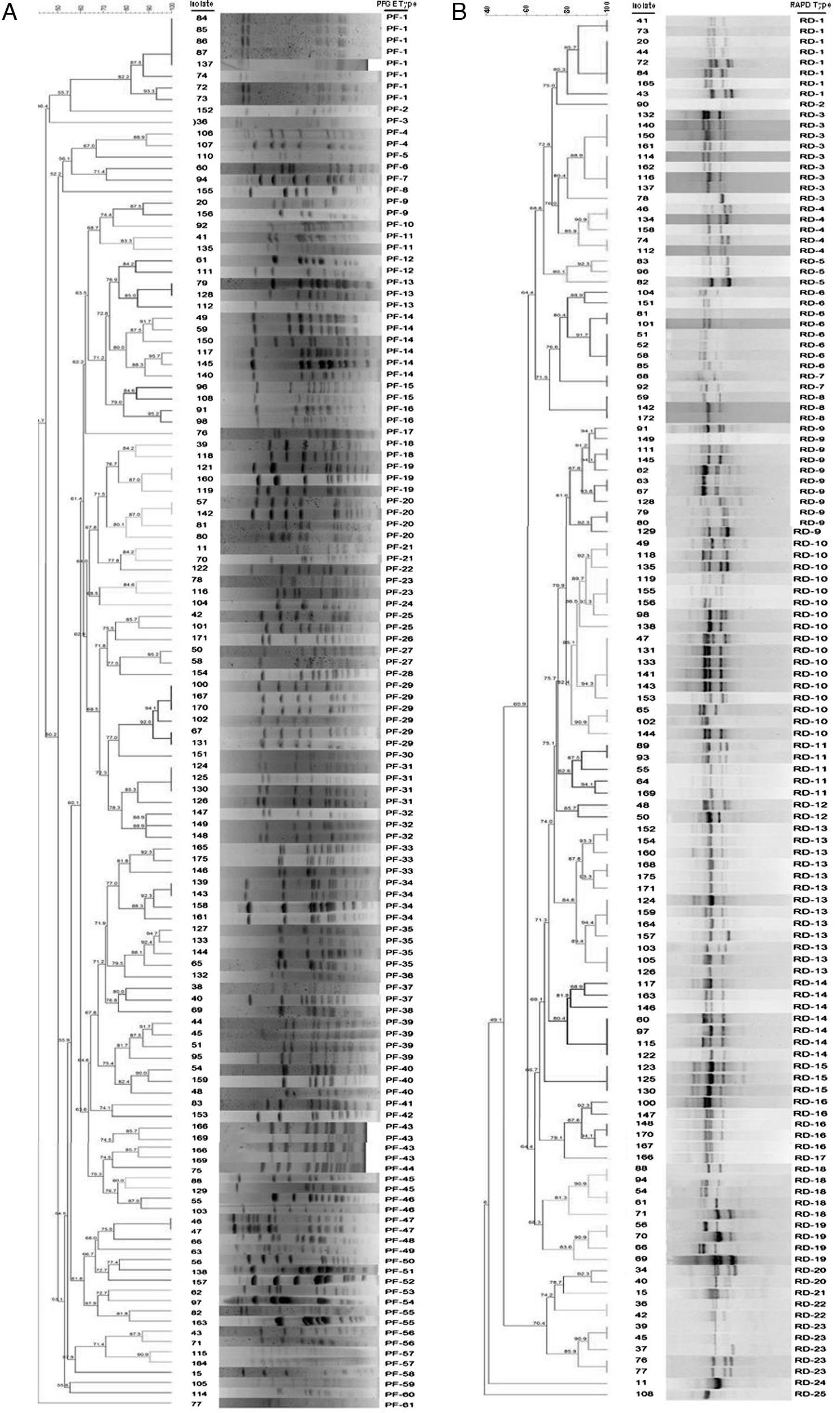
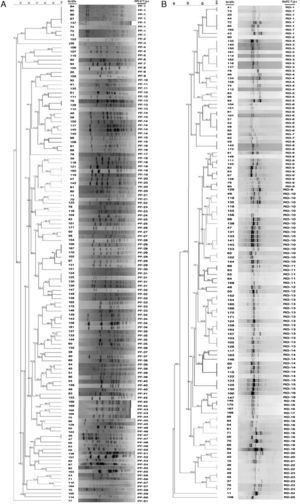
Dendrogram of pulsed-field gel electrophoresis (PFGE) (A) and random amplified polymorphic DNA (RAPD) (B) profiles of 137 Streptococcus uberis subclinical mastitis isolates collected from 35 dairy herds. Isolate code and PFGE type (A) and RAPD type (B) of each strain are also represented in the dendrogram. The dendrogram was produced by using Dice coefficients and an unweighted pair group method using arithmetic averages (UPGMA).
Forward and reverse sequences were aligned using Sequencher-DNA Sequencig Software Demo 5.1 (Gene Codes Corporation) and the consensus sequence files converted to FASTA format. Sequences were analyzed by searching the GenBank database of the National Center for Biotechnology Information via the Basic Local Alignment Search Tool (BLAST) network service. Then, the sequences were compared between them and against all existing allelic types from the database using the Codon Code Aligner software (Codon Code Corporation, Dedham, MA). The pauA sequences were also compared through the Food Microbe Tracker database at www.pathogentracker.net48.
ResultsTypability and discrimination of PFGE typingThe molecular epidemiological analysis of S. uberis isolates from milk identified by RFLP was performed on 137 isolates from 35 dairy farms. Digested chromosomal DNA generated 1–23 fragments ranging from 48.5 to 436.5kb in size, which could be resolved by PFGE (Fig. 2a). Epidemiologically related groups were identified based on a minimum 80% similarity. The analysis of the 137 isolates revealed 61 types of PFGE patterns, 29 of which were unique isolates, and 32 types, containing 2–8 isolates per type. Isolate distribution in groups was: 19 types with 2 isolates (PF 04, 9, 11, 12, 15, 16, 18, 21, 23, 25, 27, 37, 43, 45, 46, 47, 55, 56 and 57), 5 types with 3 isolates (PF 13, 19, 32, 33 and 40), 5 types with 4 isolates (PF 20, 31, 34, 35 and 39), 2 types with 6 isolates (PF14 and PF29) and 1 type with 8 isolates (PF1) (Table 1). Genotype grouping appeared to be neither related to the mastitis type (clinical or subclinical) nor to the farm origin. Epidemiologically related groups were observed among isolates from different farms. Similarities ranging between 80.1% and 100% were established in isolates retrieved from the same farm in only 11 types (PF 1, 4, 14, 19, 20, 29, 31, 32, 34, 35 and 47), suggesting a common source or horizontal transmission of the pathogen. Fifteen out of the 137 studied isolates gave a poor PFGE quality pattern; therefore, they could not be included in the analysis. These strains were classified as non-typeable, resulting in 90.5% typability for this technique according to the Epicompare software analysis.
Typability and discrimination of RAPD-PCR typingProducts obtained after RAPD-PCR amplification were 0.5–8.0kb in size (Fig. 2b). All samples exhibited at least one of these products: 1632bp, 1800bp, and 2430bp. Differences among isolates were given by products ranging between 1800 and 1632bp in size. The analysis was carried out in 137 samples using the same criteria for PFGE which allowed the identification of 25 clonal patterns. The patterns were assigned to 5 types with only one isolate and 20 types, containing 2–17 isolates. The composition of the groups was as follows: 4 types with 2 isolates (RD-7, 12, 20 and 22), 3 types with 3 isolates (RD-5, 8 and 15), 1 type with 4 isolates (RD-19), 5 types with 5 isolates (RD-4, 11, 16, 18 and 23), 1 type with 7 isolates (RD-14), 2 types with 8 isolates (RD-1 and 6), 1 with 9 isolates (RD-3), 1 with 11 isolates (RD-9), 1 with 13 isolates (RD-13), and 1 with 17 isolates (RD-10). As for PFGE, no association was found either between mastitis type or farm of origin. Thirteen isolates could not be assigned to any group given the poor quality observed in their electrophoretic pattern, and therefore they were classified as non-typeable resulting in 89% typability for this technique according to the Epicompare software analysis.
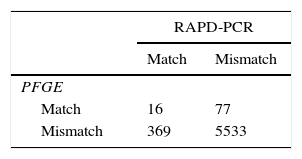
Comparison of the typing methodsTo compare the discriminatory power of PFGE and RAPD, we calculated the Simpson's diversity index for both methods (Table 2). PFGE and RAPD yielded different Simpson index values of 0.983 (95% CI 0.978–0.989) and 0.941 (95% CI 0.927–0.955), respectively. To compare the congruence between type assignments using PFGE and RAPD, we calculated the Wallace coefficients. Wallace coefficients for PFGE and RAPD indicated a weak bidirectional correspondence (0.042–0.172) between types generated by both methods. However, cross-classification of the isolates, based on matched or mismatched schemes by PFGE and RAPD, showed that the 2 typing systems were 92.6% concordant (Table 3).
PauA, encoded by the pauA gene, and SUAM, encoded by the sua gene, have been described as two of S. uberis major virulence factors and as potential vaccine immunogens against this pathogen. We studied the distribution of these genes among a population of S. uberis from our country and analyzed their sequences in 20 selected isolates (Table 1; Fig. 1). Table 1 and Fig. 1 show S. uberis isolates selected according to geographical differences and/or epidemiological dissimilarity. The encoding genes for SUAM and PauA were present in the majority of S. uberis isolates from different Argentinean dairy areas. The pauA gene was detected in 94.9% (130/137) whereas the sua gene in 97.8% (134/137).
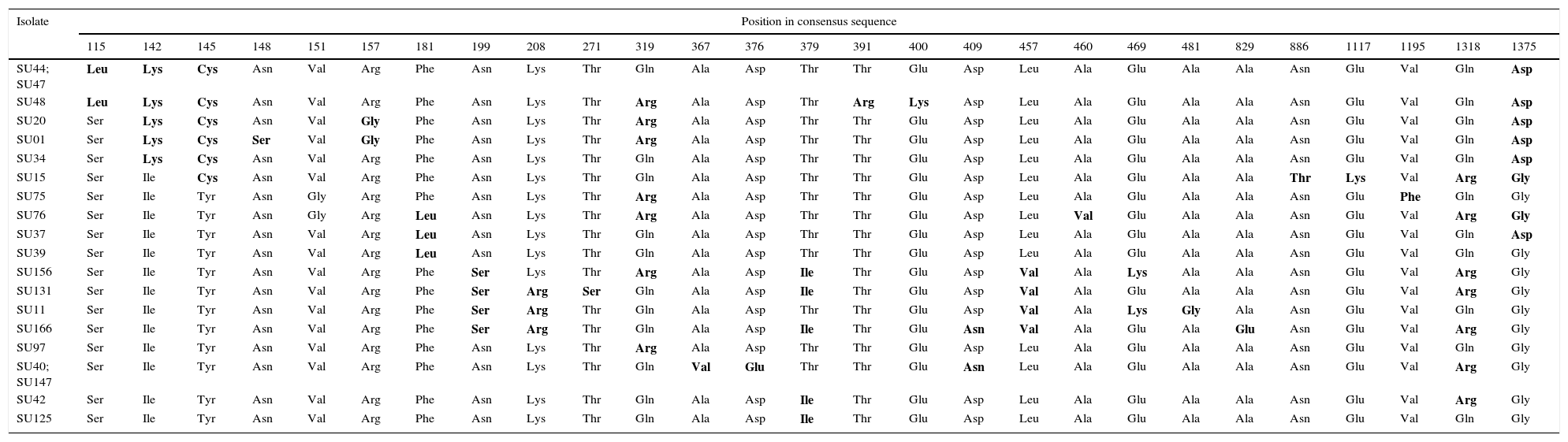
Nucleotide sequences of the sua gene from the 20 S. uberis selected isolates showed 99% identity with respect to the unique reference sequence from the GenBank (DQ232760.1)2. In concordance with the identity of nucleotide sequences, the amino acid sequences from mastitis isolates showed between 97% and 99% identity with respect to the reference sequence (GenBank ABB52003.1)2. Amino acids encoded by codons with single mutations in the sua gene are shown in Table 4. About 5–13 amino acid changes were detected in the 20 isolates. It is worth mentioning that the changes in encoded amino acids were repeated in the different isolates and suggesting conserved mutations (Table 4).
Amino acids encoded by codons with single mutations in the S. uberis adhesion molecule coding gene sua of S. uberis.
| Isolate | Position in consensus sequence | ||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 115 | 142 | 145 | 148 | 151 | 157 | 181 | 199 | 208 | 271 | 319 | 367 | 376 | 379 | 391 | 400 | 409 | 457 | 460 | 469 | 481 | 829 | 886 | 1117 | 1195 | 1318 | 1375 | |
| SU44; SU47 | Leu | Lys | Cys | Asn | Val | Arg | Phe | Asn | Lys | Thr | Gln | Ala | Asp | Thr | Thr | Glu | Asp | Leu | Ala | Glu | Ala | Ala | Asn | Glu | Val | Gln | Asp |
| SU48 | Leu | Lys | Cys | Asn | Val | Arg | Phe | Asn | Lys | Thr | Arg | Ala | Asp | Thr | Arg | Lys | Asp | Leu | Ala | Glu | Ala | Ala | Asn | Glu | Val | Gln | Asp |
| SU20 | Ser | Lys | Cys | Asn | Val | Gly | Phe | Asn | Lys | Thr | Arg | Ala | Asp | Thr | Thr | Glu | Asp | Leu | Ala | Glu | Ala | Ala | Asn | Glu | Val | Gln | Asp |
| SU01 | Ser | Lys | Cys | Ser | Val | Gly | Phe | Asn | Lys | Thr | Arg | Ala | Asp | Thr | Thr | Glu | Asp | Leu | Ala | Glu | Ala | Ala | Asn | Glu | Val | Gln | Asp |
| SU34 | Ser | Lys | Cys | Asn | Val | Arg | Phe | Asn | Lys | Thr | Gln | Ala | Asp | Thr | Thr | Glu | Asp | Leu | Ala | Glu | Ala | Ala | Asn | Glu | Val | Gln | Asp |
| SU15 | Ser | Ile | Cys | Asn | Val | Arg | Phe | Asn | Lys | Thr | Gln | Ala | Asp | Thr | Thr | Glu | Asp | Leu | Ala | Glu | Ala | Ala | Thr | Lys | Val | Arg | Gly |
| SU75 | Ser | Ile | Tyr | Asn | Gly | Arg | Phe | Asn | Lys | Thr | Arg | Ala | Asp | Thr | Thr | Glu | Asp | Leu | Ala | Glu | Ala | Ala | Asn | Glu | Phe | Gln | Gly |
| SU76 | Ser | Ile | Tyr | Asn | Gly | Arg | Leu | Asn | Lys | Thr | Arg | Ala | Asp | Thr | Thr | Glu | Asp | Leu | Val | Glu | Ala | Ala | Asn | Glu | Val | Arg | Gly |
| SU37 | Ser | Ile | Tyr | Asn | Val | Arg | Leu | Asn | Lys | Thr | Gln | Ala | Asp | Thr | Thr | Glu | Asp | Leu | Ala | Glu | Ala | Ala | Asn | Glu | Val | Gln | Asp |
| SU39 | Ser | Ile | Tyr | Asn | Val | Arg | Leu | Asn | Lys | Thr | Gln | Ala | Asp | Thr | Thr | Glu | Asp | Leu | Ala | Glu | Ala | Ala | Asn | Glu | Val | Gln | Gly |
| SU156 | Ser | Ile | Tyr | Asn | Val | Arg | Phe | Ser | Lys | Thr | Arg | Ala | Asp | Ile | Thr | Glu | Asp | Val | Ala | Lys | Ala | Ala | Asn | Glu | Val | Arg | Gly |
| SU131 | Ser | Ile | Tyr | Asn | Val | Arg | Phe | Ser | Arg | Ser | Gln | Ala | Asp | Ile | Thr | Glu | Asp | Val | Ala | Glu | Ala | Ala | Asn | Glu | Val | Arg | Gly |
| SU11 | Ser | Ile | Tyr | Asn | Val | Arg | Phe | Ser | Arg | Thr | Gln | Ala | Asp | Thr | Thr | Glu | Asp | Val | Ala | Lys | Gly | Ala | Asn | Glu | Val | Gln | Gly |
| SU166 | Ser | Ile | Tyr | Asn | Val | Arg | Phe | Ser | Arg | Thr | Gln | Ala | Asp | Ile | Thr | Glu | Asn | Val | Ala | Glu | Ala | Glu | Asn | Glu | Val | Arg | Gly |
| SU97 | Ser | Ile | Tyr | Asn | Val | Arg | Phe | Asn | Lys | Thr | Arg | Ala | Asp | Thr | Thr | Glu | Asp | Leu | Ala | Glu | Ala | Ala | Asn | Glu | Val | Gln | Gly |
| SU40; SU147 | Ser | Ile | Tyr | Asn | Val | Arg | Phe | Asn | Lys | Thr | Gln | Val | Glu | Thr | Thr | Glu | Asn | Leu | Ala | Glu | Ala | Ala | Asn | Glu | Val | Arg | Gly |
| SU42 | Ser | Ile | Tyr | Asn | Val | Arg | Phe | Asn | Lys | Thr | Gln | Ala | Asp | Ile | Thr | Glu | Asp | Leu | Ala | Glu | Ala | Ala | Asn | Glu | Val | Arg | Gly |
| SU125 | Ser | Ile | Tyr | Asn | Val | Arg | Phe | Asn | Lys | Thr | Gln | Ala | Asp | Ile | Thr | Glu | Asp | Leu | Ala | Glu | Ala | Ala | Asn | Glu | Val | Gln | Gly |
| Isolate | Position in consensus sequence | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1390 | 1633 | 1717 | 1786 | 1795 | 1804 | 1873 | 2005 | 2008 | 2023 | 2080 | 2104 | 2195 | 2395 | 2422 | 2455 | 2458 | 2464 | 2477 | 2506 | 2512 | 2524 | 2545 | 2749 | Total Mutations | |
| SU44; SU47 | Thr | Pro | Ala | Ile | Ala | Ala | Ile | Ala | Arg | Asn | Leu | Asn | Lys | Asn | Arg | Glu | Val | Asp | Glu | Val | Glu | Thr | Val | Ile | 9 |
| SU48 | Thr | Pro | Ala | Ile | Ala | Ala | Ile | Ala | Arg | Asn | Leu | Asn | Lys | Asn | Arg | Glu | Val | Asp | Glu | Val | Glu | Thr | Val | Ile | 12 |
| SU20 | Thr | Pro | Ala | Ile | Ala | Ala | Ile | Ala | Arg | Asn | Leu | Asn | Lys | Asn | Arg | Glu | Val | Asp | Glu | Val | Glu | Thr | Val | Ile | 10 |
| SU01 | Thr | Pro | Ala | Ile | Ala | Ala | Ile | Ala | Arg | Asn | Leu | Asn | Lys | Asn | Arg | Glu | Val | Asp | Glu | Val | Glu | Thr | Val | Ile | 11 |
| SU34 | Thr | Pro | Ala | Ile | Ala | Ala | Ile | Ala | Arg | Asn | Leu | Asn | Lys | Asn | Arg | Glu | Val | Asp | Glu | Val | Glu | Thr | Val | Ile | 8 |
| SU15 | Ile | Pro | Pro | Thr | Ala | Ala | Ile | Ala | Leu | Asp | Leu | Ser | Arg | Asn | Arg | Glu | Ala | Asp | Glu | Val | Lys | Thr | Ile | Val | 10 |
| SU75 | Thr | Pro | Ala | Ile | Ala | Ala | Ile | Ala | Arg | Asn | Leu | Asn | Lys | Asn | Arg | Glu | Val | Asp | Glu | Val | Lys | Thr | Ile | Ile | 8 |
| SU76 | Ile | Pro | Ala | Thr | Ala | Ala | Ile | Ala | Leu | Asp | Phe | Ser | Lys | Ser | Arg | Lys | Ala | Asp | Glu | Met | Glu | Ala | Val | Ile | 11 |
| SU37 | Thr | Pro | Ala | Ile | Ala | Ala | Ile | Ala | Arg | Asn | Leu | Asn | Lys | Asn | Arg | Glu | Val | Asp | Glu | Val | Glu | Thr | Val | Ile | 7 |
| SU39 | Ile | Pro | Ala | Thr | Ala | Thr | Val | Ser | Leu | Asp | Leu | Ser | Lys | Ser | Arg | Lys | Ala | Asp | Glu | Met | Glu | Thr | Val | Ile | 8 |
| SU156 | Ile | Ser | Ala | Thr | Ala | Ala | Ile | Ala | Leu | Asp | Phe | Ser | Lys | Ser | Lys | Lys | Ala | Asp | Ala | Val | Glu | Thr | Val | Ile | 13 |
| SU131 | Ile | Pro | Ala | Thr | Ala | Ala | Ile | Ala | Leu | Asp | Phe | Ser | Lys | Ser | Lys | Lys | Ala | Asp | Ala | Val | Glu | Thr | Val | Ile | 12 |
| SU11 | Ile | Pro | Ala | Thr | Val | Ala | Ile | Ala | Leu | Asp | Leu | Ser | Lys | Asn | Arg | Lys | Ala | Ala | Glu | Val | Glu | Thr | Val | Ile | 9 |
| SU166 | Ile | Pro | Ala | Thr | Ala | Ala | Ile | Ala | Leu | Asp | Leu | Ser | Lys | Ser | Lys | Lys | Ala | Asp | Glu | Val | Glu | Thr | Val | Ile | 11 |
| SU97 | Ile | Pro | Ala | Thr | Val | Ala | Ile | Ala | Leu | Asp | Leu | Ser | Lys | Asn | Arg | Lys | Ala | Ala | Glu | Val | Glu | Thr | Val | Ile | 5 |
| SU40; SU147 | Ile | Pro | Ala | Thr | Ala | Ala | Ile | Ala | Leu | Asp | Leu | Ser | Lys | Asn | Arg | Glu | Val | Asp | Glu | Val | Lys | Thr | Ile | Ile | 6 |
| SU42 | Ile | Pro | Ala | Thr | Ala | Ala | Ile | Ala | Leu | Asp | Phe | Ser | Lys | Ser | Lys | Lys | Ala | Asp | Ala | Val | Glu | Thr | Val | Ile | 8 |
| SU125 | Thr | Pro | Ala | Ile | Ala | Ala | Ile | Ala | Arg | Asn | Leu | Asn | Lys | Asn | Arg | Glu | Val | Asp | Glu | Val | Lys | Thr | Ile | Ile | 8 |
The difference in amino acid sequence from the majority are bolded.
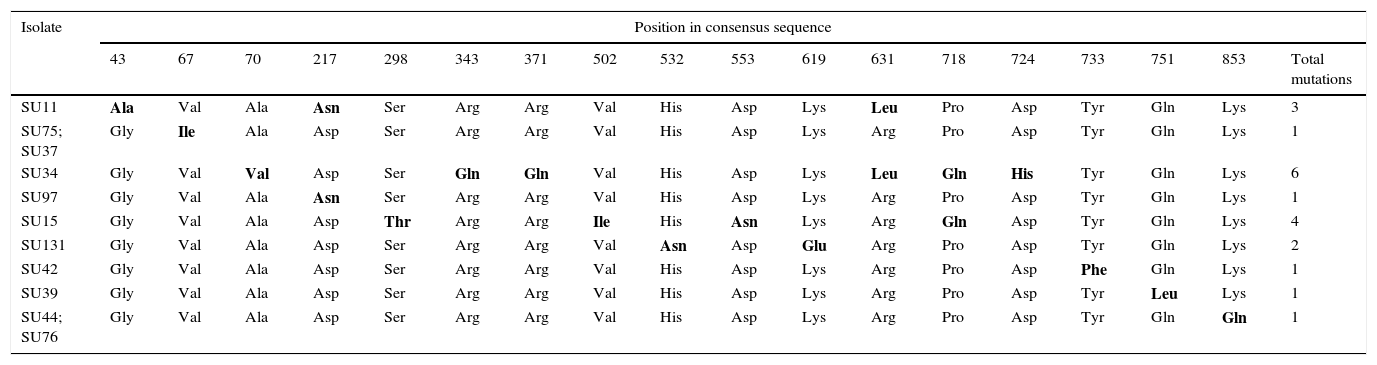
Nucleotide sequences of the pauA gene from the 20 selected S. uberis mastitis isolates showed between 95% and 100% identity with respect to all the 55 sequences registered in GenBank and to 30 sequences registered in the Food Microbe Tracker database. Similarly, the amino acid sequences showed 96–100% identity with respect to the reference sequences. Amino acids encoded by codons with single mutations in pauA are shown in Table 5. About 1–6 mutations that resulted in amino acid change were detected in 11 isolates. Sequence data of the sua and pauA genes were registered in GenBank under accession numbers KT006548–KT006587.
Amino acids encoded by codons with single mutations in the plasminogen activator coding gene pauA of S. uberis.
| Isolate | Position in consensus sequence | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 43 | 67 | 70 | 217 | 298 | 343 | 371 | 502 | 532 | 553 | 619 | 631 | 718 | 724 | 733 | 751 | 853 | Total mutations | |
| SU11 | Ala | Val | Ala | Asn | Ser | Arg | Arg | Val | His | Asp | Lys | Leu | Pro | Asp | Tyr | Gln | Lys | 3 |
| SU75; SU37 | Gly | Ile | Ala | Asp | Ser | Arg | Arg | Val | His | Asp | Lys | Arg | Pro | Asp | Tyr | Gln | Lys | 1 |
| SU34 | Gly | Val | Val | Asp | Ser | Gln | Gln | Val | His | Asp | Lys | Leu | Gln | His | Tyr | Gln | Lys | 6 |
| SU97 | Gly | Val | Ala | Asn | Ser | Arg | Arg | Val | His | Asp | Lys | Arg | Pro | Asp | Tyr | Gln | Lys | 1 |
| SU15 | Gly | Val | Ala | Asp | Thr | Arg | Arg | Ile | His | Asn | Lys | Arg | Gln | Asp | Tyr | Gln | Lys | 4 |
| SU131 | Gly | Val | Ala | Asp | Ser | Arg | Arg | Val | Asn | Asp | Glu | Arg | Pro | Asp | Tyr | Gln | Lys | 2 |
| SU42 | Gly | Val | Ala | Asp | Ser | Arg | Arg | Val | His | Asp | Lys | Arg | Pro | Asp | Phe | Gln | Lys | 1 |
| SU39 | Gly | Val | Ala | Asp | Ser | Arg | Arg | Val | His | Asp | Lys | Arg | Pro | Asp | Tyr | Leu | Lys | 1 |
| SU44; SU76 | Gly | Val | Ala | Asp | Ser | Arg | Arg | Val | His | Asp | Lys | Arg | Pro | Asp | Tyr | Gln | Gln | 1 |
The difference in amino acid sequence from the majority are bolded.
S. uberis is a well-known pathogen causing bovine IMI worldwide. However, there is scant epidemiological information on S. uberis isolated from bovine milk in Argentina19. In the present study, PFGE and RAPD were used for the molecular characterization of S. uberis isolated from cows belonging to herds located in the main dairy areas of Argentina. Based of this information a reliable representative set of isolates was selected for the study of the pauA and sua genes.
As shown in previous studies1,32,36,43, PFGE has proven to be a highly discriminatory method. In the present study, PFGE was able to resolve many isolates that were indistinguishable by RAPD-PCR. Some isolates from different herds that were defined as belonging to the same type (100% similarities) were associated with different types of mastitis (both clinical and subclinical), in agreement with previous studies15,29. The PFGE patterns for S. uberis showed great variation; among the 137 isolates (collected from 122 cows on 35 farms), 61 distinct PFGE profiles were observed. This high level of heterogeneity is in accordance with classical epidemiological studies from Argentina and other countries1,19,32,42. However, we found eleven cases of one type isolated from different animals in the same dairy herd. This finding suggests that cows were infected by the same organism, either from a common source or by spread from one quarter or cow to another. Poor milking hygiene and faulty milking machine functioning could contribute to S. uberis transmission among cows47. The total epidemiological data reported in this work allows to consider S. uberis as an environmental opportunistic pathogen, with a limited number of dominant types and a great variety of strains.
RAPD typing relies on non-stringent reaction conditions for the amplification of arbitrary target sites26. Low stringent reaction conditions are associated with difficulty to achieve high pattern repeatability10. Target DNA concentration conditions and the thermal cycling program were standardized in our fingerprinting method to yield constant and reproducible results. The analysis of the 137 isolates distinguished 25 distinct RAPD profiles. Conserved band pattern obtained with RAPD-PCR disagreed with those previously reported by groups from New Zealand and United States11,45. These discrepancies indicate that the method does not allow to compare results from different geographical origins. However, RAPD allows to perform epidemiological studies in less time at lower costs45.
The comparative analysis of RAPD and PFGE results indicated that isolates belonging to the same type according to RAPD exhibited different PFGE patterns. The discriminatory power and congruence between both methods were compared using the Simpson's index of diversity and Wallace's coefficients. Our results indicated that both typing methods had a high discriminatory power between epidemiologically non-related isolates as indicated by the Simpson's index of diversity. Although the Simpson's diversity index of the FPGE method was higher than that of the RAPD method, the difference was modest. The congruence between types defined by PFGE and RAPD yielded low values, as reflected by the Wallace coefficients (0.042 and 0.172), indicating a weak bidirectional agreement among types generated by both methods. PFGE is a technique of choice for short-term or local epidemiological studies, since even minor genetic changes can lead to a three-fragment difference in the PFGE banding pattern1. Moreover, several studies have shown that PFGE has greater discriminatory power than MLST32,38,40. Gillespie and Oliver12 compared RAPD and PFGE for differentiation of S. uberis isolates, concluding that PFGE had higher discriminatory power than RAPD on the basis of the number of groups that could be differentiated by each method. Similar results were obtained in the present study. However, considering the high concordance between both methods (96.2%), we can conclude that any given pair of isolates distinguished by one method tended to be distinguished by the other. To our knowledge, this is the first study reporting a statistical analysis for comparison of these two techniques applied for S. uberis molecular epidemiology.
A successful vaccine should confer broad protection against a multitude of strains. This approach requires a detailed knowledge of the epidemiology and pathogenesis of the organism. Despite the severe economic impact caused by this infection, the virulence factors associated with S. uberis pathogenesis are not well understood2. Plasminogen activators such as PauA have been proposed as important intermediaries to obtain nutrients for optimal growth of the organism44. In addition, the successful establishment of IMI depends on adherence, internalization, and intracellular persistence28. The SUAM was found to play a central role in S. uberis adherence to bovine mammary epithelial cells8, contributing to infection persistence. These two virulence factors have been previously studied as potential immunogens3,21. However, it is unknown whether genetic variability could have modified the effectiveness they demonstrated in preclinical trials. In the present study, 94.9% of the strains studied carried the pauA gene. The prevalence of the pauA gene was previously reported as ranging from 61.5% in Argentina to 100% in India17,33,36. To get further insight, we studied the nucleotide and amino acid sequences from 20 selected S. uberis mastitis isolates. We observed an identity greater than 95% with respect to all the 55 sequences registered in GenBank and to the 30 sequences registered in the Food Microbe Tracker database. Furthermore, amino acid sequences showed a low number of mutations. Mutations at positions 43, 67, 217, 343, 371, 631, 718 and 724 of the consensus sequences coincided with those described by Zadoks et al47. In fact, the mutations observed in other positions found in this work were not described until now. Therefore, pauA is present in most isolates and interestingly, it is a highly conserved gene. Correspondingly, the presence of the sua gene was detected in 97.8% of the 137 isolates. Previous works reported a prevalence of the sua gene of 83.3%33 in Argentina, 84.6% in India36 and 100% in New Zealand, United States and England23,46. In addition, the nucleotide and amino acid sequences from the 20 selected S. uberis mastitis isolates had an identity greater than 97% with respect to the reference sequence (ABB52003.1). The study of amino acid sequences showed several repeated mutations in different isolates. Yuan et al. described similar results in sua gene conservation among isolates from different countries46. The relevance of the present findings, compared with previous studies, relies on the higher number (n=137), the strict criteria used for the selection and broad distribution of the isolates analyzed. All together these results demonstrate the high prevalence and conservation of the sua and pauA genes in Argentinean S. uberis isolates, which was also reflected in their amino acid sequence analysis. To our knowledge, this is the first study that compared these two virulence factors in sequences from field isolates versus reported GenBank sequences from different countries.
It has been suggested that the genetic clonal diversity of S. uberis is an obstacle for the development of an effective vaccine since a broadly-reactive immunogen with field strains has not been obtained so far9,21. However, previous works and the present study demonstrated that S. uberis isolates belonging to different PFGE profiles showed conserved gene sequences32. In addition, it has to be taken in account that the minimal variation observed in gene carriage highlights a potential problem with respect to the development of subunit vaccines against S. uberis, since vaccines based on a single antigen may not provide protection against all strains of the pathogen even using conserved genes. Thus, we found that those isolates that did not carry the sua gene harbor the pauA gene and vice versa (Table 1), and only in two isolates (1.45%) none of these genes were detected. All together, our results showed that the pauA and sua genes are conserved across different geographical areas and are present in most S. uberis isolates despite the high genetic variability observed, which provides further support for the use of these virulence factors as potential vaccine components against S. uberis.
Ethical disclosuresProtection of human and animal subjectsThe authors declare that the procedures followed were in accordance with the regulations of the relevant clinical research ethics committee and with those of the Code of Ethics of the World Medical Association (Declaration of Helsinki).
Confidentiality of dataThe authors declare that no patient data appear in this article.
Right to privacy and informed consentThe authors declare that no patient data appear in this article.
FundingThis work was supported by grants from FONCyT (M.S.B., PICT Bicentenario 2010-1309), Q5 (F.R.B., PICT Bicentenario 2010-0733), (L.F.C. and I.S.M., PICT 1175) and from Fundación Banco de la Provincia de Santa Fe (M.S.B.).
Conflict of interestThe authors declare that they have no conflicts of interest.
We thank M.V. Liliana Tirante (Lactodiagnóstico Sur, Olivos, Buenos Aires) for providing the isolates and Dr. Andrea Puebla (Instituto de Biotecnología, CICVyA Castelar, INTA) for sequencing service. M.S.P. is a fellow from CONICET, I.S.M. and F.R.B. are independent and adjunct members of the research career of CONICET.