Helicobacter pylori is a gastric pathogen that is widely recognized as a causative agent of gastric disease. Its eradication is variable, mainly due to increased resistance to clarithromycin. Our objective was: to evaluate (i) if the biopsy specimen used for the rapid urease test is a useful sample to detect resistance to clarithromycin by PCR-RFLP and (ii) the distribution of A2142G and A2143G point mutations in the 23S rRNA gene, in relation to virulence factors in our region. Gastric specimens were collected from adult dyspeptic patients (n=141) and H. pylori was investigated by the rapid urease test, histopathological analysis and PCR for the hsp60 gene. Clarithromycin resistance was detected by PCR-RFLP in 62 H. pylori (+) paired biopsy specimens submitted to molecular analysis and the rapid urease test. H. pylori virulence factors were analyzed by multiplex PCR using specific primers for the cagA, vacA and babA2 genes. Thirteen out of 62 strains (20.9%) were resistant to clarithromycin: 6/13 (46.2%) harbored the A2143G mutation whereas 7/13 (53.8%) carried the A2142G point mutation. vacA m1s1 was the most frequent genotype among the resistant strains. In conclusion, the biopsy specimens used for the rapid urease test were suitable samples for clarithromycin resistance detection in patients infected with H. pylori, which became especially useful in cases where the number or size of the biopsies is limited. In addition, this is the first report of a molecular analysis for clarithromycin resistance performed directly from gastric biopsies in our region.
Helicobacter pylori es un patógeno ampliamente reconocido como causante de enfermedad gástrica. Su erradicación es variable, principalmente debido al incremento de la resistencia a claritromicina. Nuestros objetivos fueron evaluar la utilidad de la biopsia usada para realizar el test rápido de ureasa en la detección de resistencia a claritromicina por PCR-RFLP y conocer la distribución de las mutaciones puntuales A2142G y A2143G en el gen ARNr 23S, en relación con los factores de virulencia en nuestra región. Se recolectaron muestras gástricas (n=141) provenientes de pacientes adultos dispépticos y se investigó la presencia de H. pylori mediante el test rápido de ureasa, análisis histopatológico y PCR para el gen hsp60. La resistencia a claritromicina se analizó por PCR-RFLP en 62 muestras pareadas de biopsias gástricas H. pylori+ destinadas al análisis molecular y al test rápido de ureasa. Los factores de virulencia de H. pylori fueron analizados mediante PCR multiplex usando oligonucleótidos específicos para los genes cagA, vacA y babA2. Trece de 62 cepas (20,9%) fueron resistentes a claritromicina, 6/13 (46,2%) llevaron la mutación A2143G, mientras que 7/13 (53,8%) presentaron la mutación A2142G. El genotipo vacA s1m1 fue el más frecuente entre las cepas resistentes a claritromicina. En conclusión, las biopsias destinadas al test rápido de ureasa fueron muestras apropiadas para la detección de la resistencia a claritromicina en pacientes infectados con H. pylori. Esto es especialmente útil en aquellos casos en los que el número o el tamaño de las muestras son limitados. Además, este es el primer reporte de estudio de resistencia a claritromicina (mediante técnicas moleculares), directamente de biopsias gástricas en nuestra región.
Helicobacter pylori is an etiological agent for several gastroduodenal diseases with various clinical manifestations ranging from dyspepsia, chronic gastritis, gastric atrophy and peptic ulcer disease to gastric adenocarcinoma and primary gastric B-cell lymphoma. The different clinical presentations may be due to specific virulence factors of the bacterium (among others, cagA, vacA and babA genes) as well as intrinsic host factors. Furthermore, it has been classified as a Class I carcinogen by the International Agency for Research on Cancer (IARC)27 as well as by the World Health Organization. H. pylori is known to colonize the gastric mucosa in 50% of the human population worldwide, with varying prevalence rates among different geographical regions, with the highest rates observed in developing countries13,27. In this regard, the detection of H. pylori in gastric biopsies includes bacterial culture, the rapid urease test and molecular analysis by nucleic acid amplification.
Early eradication of H. pylori has been proven to reduce the incidence rate of gastric cancer and improve ulcer healing27. Nowadays, the most widely used treatment regimens include the standard triple therapy, which comprises two out of three antibiotics, clarithromycin and amoxicillin or metronidazole, together with a proton pump inhibitor (PPI)16. The efficacy of this therapy has dramatically declined over the last decade, mainly due to increasing clarithromycin resistance rates observed in many regions of the world21. In Argentina there are few reports about clarithromycin resistance; however, current evidence indicates that such resistance is increasing24,29–31. Furthermore, as reported by Zerbetto de Palma et al.31, such clarithromycin-resistance rate increase is tied to an increased rate in multidrug resistance.
Clarithromycin activity depends on the capacity to inhibit protein synthesis by binding to the bacterial 50S ribosomal subunit and point mutations in the peptidyl transferase region encoded in the domain V of 23S rRNA are responsible for macrolide resistance by inhibition of the binding between the antibiotic and the ribosomal unit. The most frequent mutations associated with clarithromycin resistance are A2143G and A2142G in the 23S rRNA gene and, to a lesser extent, A2142C26.
Phenotypic and genotypic resistance to clarithromycin can be assessed by culture (agar diffusion, agar dilution, broth microdilution, E-test) and PCR-based techniques, respectively. Because of the slow growth and particular requirements of H. pylori culture, phenotypic antimicrobial susceptibility tests are not routinely performed in clinical practice. For this reason, molecular tests targeting H. pylori resistance-associated gene mutations appear as a useful alternative for the detection of clarithromycin resistance. Taking into account the above mentioned data, the aims of this study were to evaluate: (i) if the biopsy samples used for the rapid urease test is a useful sample to detect resistance to clarithromycin by PCR-RFLP and (ii) the distribution of A2142G and A2143G point mutations in relation to virulence factors in our region.
Materials and methodsAdult dyspeptic patients (n=141) attending the Gastroenterology Service of “Dr. José María Cullen” Hospital (Santa Fe, Argentina) from February 2014 to March 2015 were enrolled in the study, after informed consent was obtained. All of them underwent upper gastrointestinal endoscopy. Patients who had received antibiotic therapy and/or PPIs in the last 15 days were excluded from the study.
H. pylori detection: gastric specimens were collected according to the modified Sydney Protocol. H. pylori was investigated by the rapid urease test, histopathological analysis and PCR for the hsp60 gene.
DNA from patients whose samples were H. pylori-positive by the histopathological test and PCR amplification of the hsp60 gene12 were included for resistance testing and for virulence factors analysis. DNA extraction was carried out as follows: biopsy samples for the rapid urease test as well as the molecular analysis were collected in Christensen's urea broth and in saline solution, respectively; in the molecular diagnostics laboratory, solutions were discarded; 400μl TES and 2μl of 20mg/ml proteinase K were added and tubes were incubated for 2h at 60°C (or overnight at 4°C). Then, 125μl of 5M NaCl were added. After mixing, 400μl of chloroform were added and tubes were vigorously shaken until a milky solution formed. Tubes were centrifuged at 10000×g for 3min and the upper aqueous phase was transferred to a new tube (600μl). Equal volume (600μl) of cold 95% ethanol (−20°C) was added and tubes were inverted 3 times to condense the DNA. A centrifugation at 10000×g for 2min was carried out and the supernatant was removed. The pellet was washed with 1ml cold 70% ethanol (−20°C) and each tube was centrifuged for 1min at 10000×g. After discarding the supernatant, the DNA was dried by placing the tube upside down on absorbent paper. Finally, the dried pellet was resuspended in 50–200μl of TE 10:1 pH 8.0 (with vortex) and incubated at 56°C until complete dissolution.
PCR-RFLP (clarithromycin resistance detection). PCR was performed using the primers described by Suzuki et al.26 which amplified a fragment of 768pb corresponding to the domain V of the 23S rRNA gene: Hp23Sr6 sense (5′ -CACACAGGTAGATGAGATGAGTA-3′) and Hp23Sr7 antisense (5′ -CACACAGAACCACCGGATCACTA-3′). An individual PCR reaction (final volume: 25μl) was done by a mix consisting of 12.5μl of 2X GoTaq Green MMix (Promega, USA), 1μl of forward/reverse primers mix (10μM), 2.5μl of DNA and 9μl nuclease-free water. PCR conditions were: 94°C 5min followed by 40 cycles of denaturation at 94°C for 30s, annealing at 50°C for 30s and extension at 72°C for 30s and one cycle at 72°C during 7min. Two aliquots of the amplification reactions (10μl) were digested separately with MboII and BsaI (New England BioLabs) for 1h at 37°C. These enzymes identified mutations in the H. pylori domain V of the 23S rRNA gene at positions 2142 and 2143, respectively.
Virulence factor detection. The cagA, babA2 and vacA genes were analyzed by multiplex PCR using sequence specific primers (0.4μM each one) and GoTaq® Green Master Mix, (Promega Corp) (25μl/tube)8,10,12. PCR cycling conditions were: 94°C for 5min followed by 40 cycles of denaturation at 94°C for 30s, annealing at 50°C for 30s and extension at 72°C for 30s. Finally, a cycle at 72°C for 7min was carried out. In all cases, PCR products were visualized after electrophoresis in 2% agarose gel stained with Sybr Safe (Life Technologies, USA).
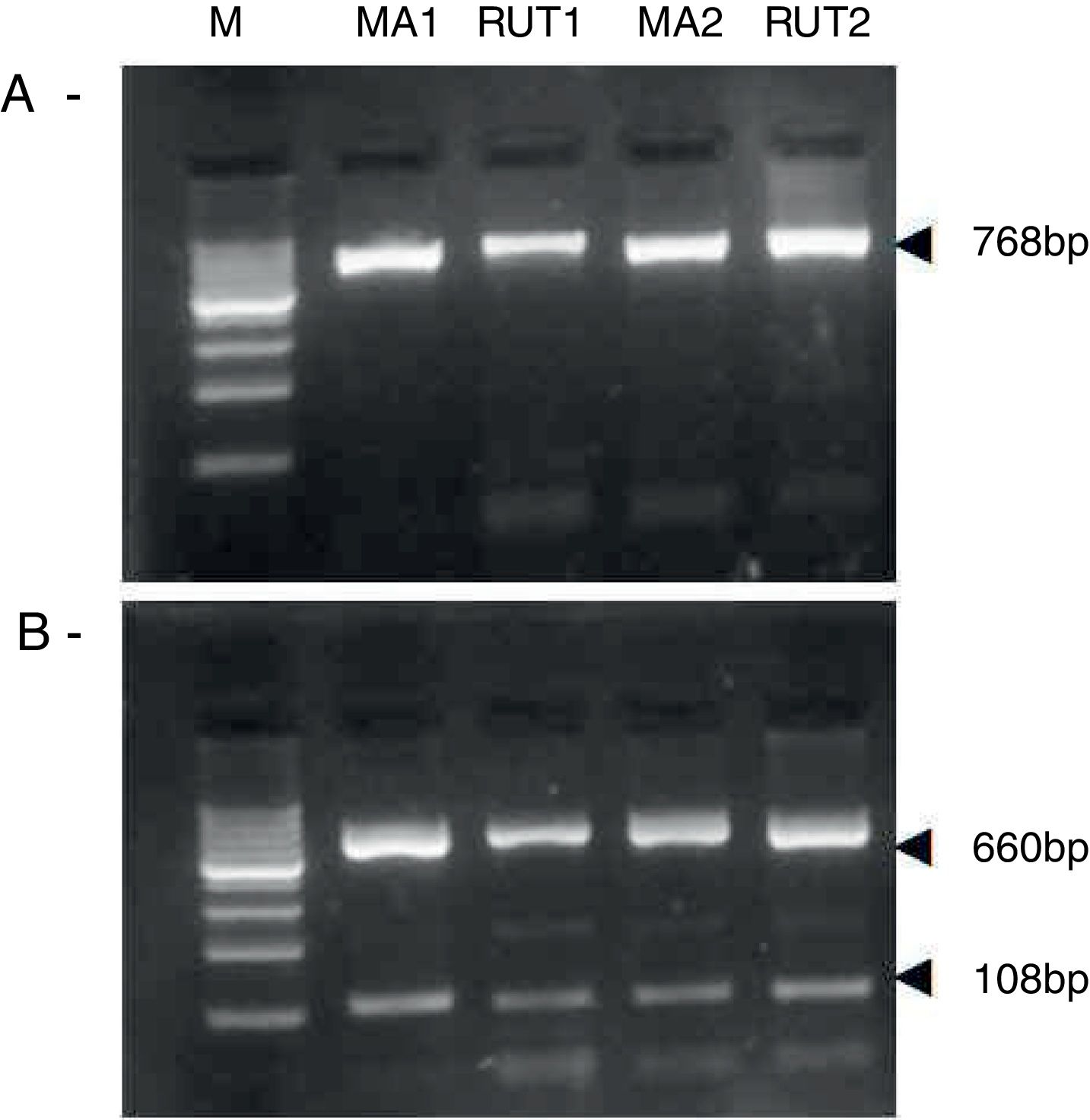
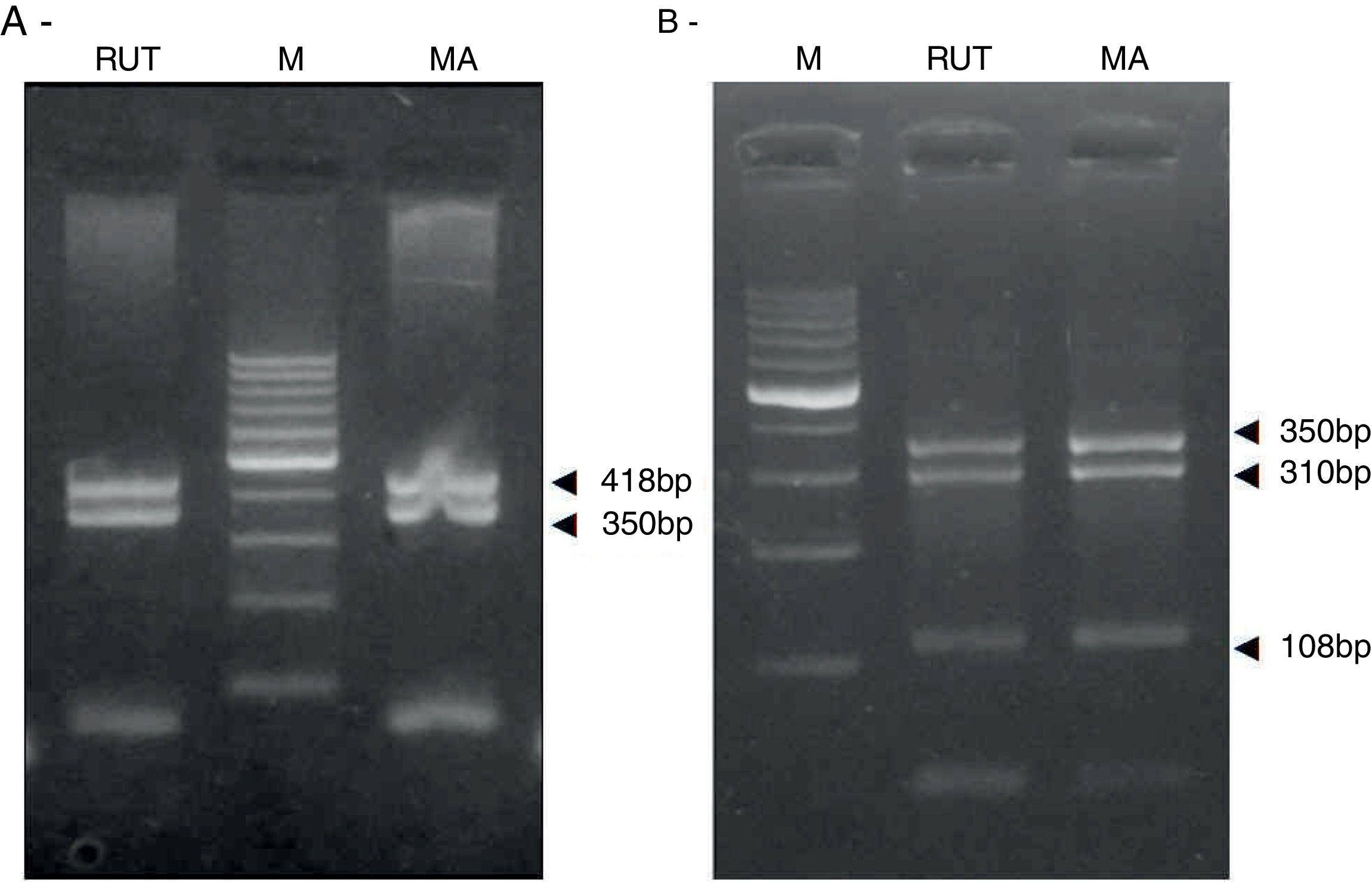
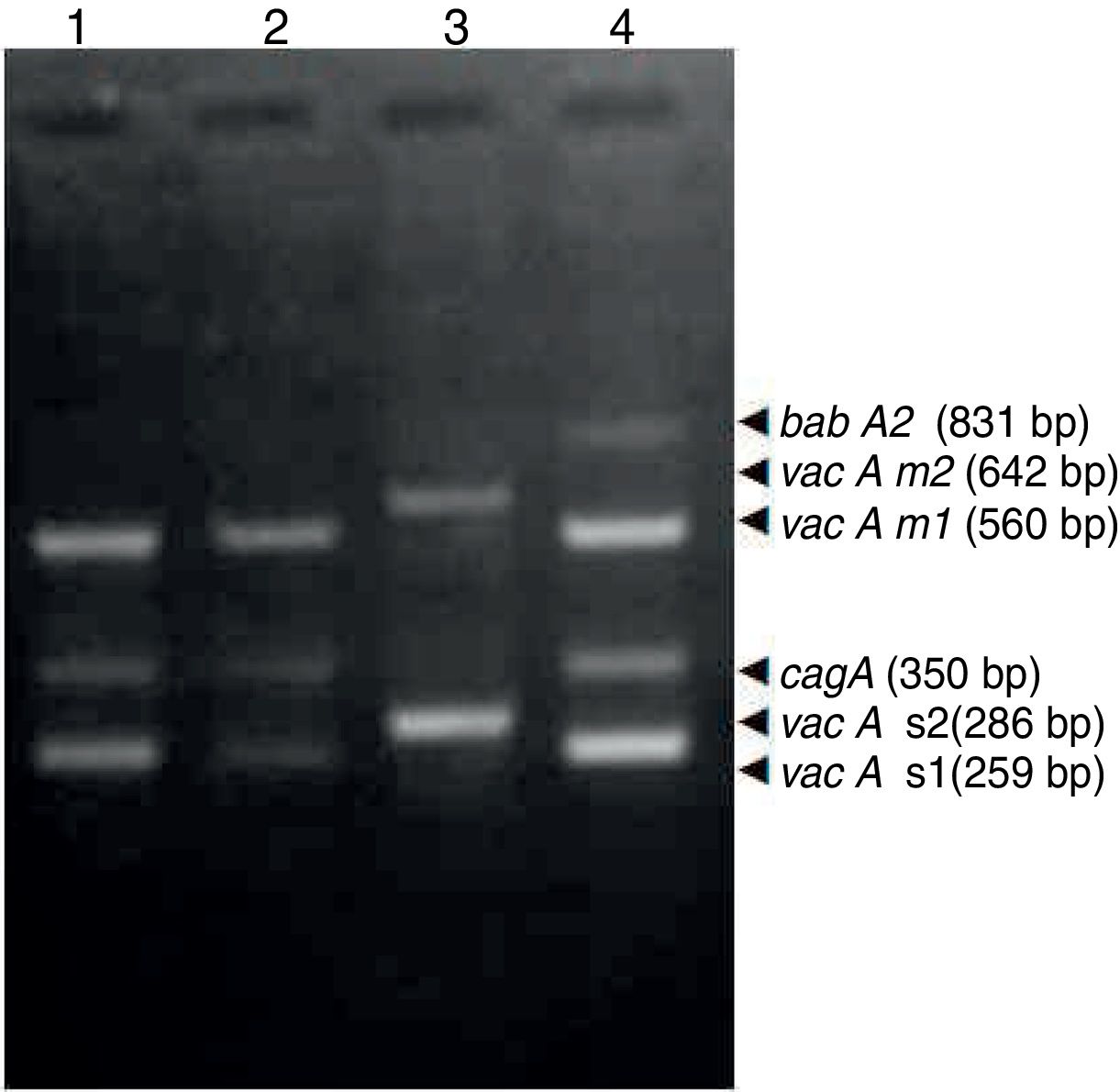
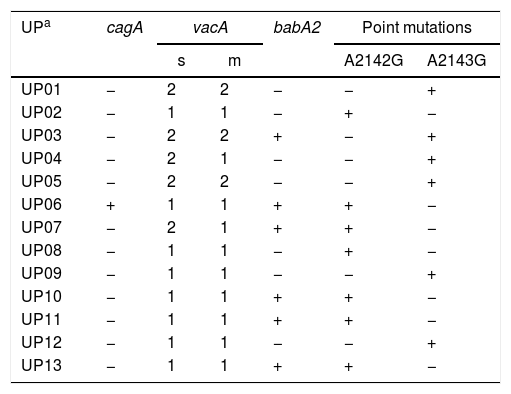
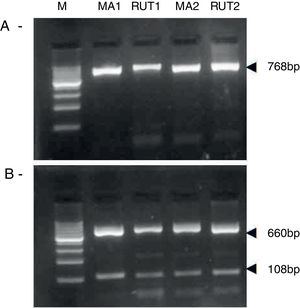
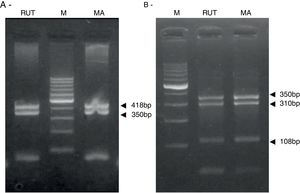
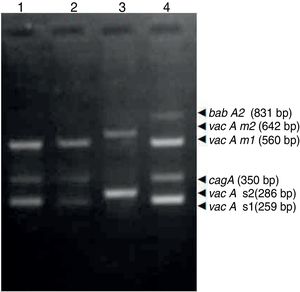
ResultsBiopsies of 62 adult patients infected with H. pylori (as assessed by PCR and histology) were analyzed by PCR-RFLP for the clarithromycin-resistance analysis. The test was performed simultaneously on biopsy samples destined to molecular analysis as well as those for the rapid urease test. In both samples, a 768-bp product can be specifically amplified with similar yield. In order to investigate the presence of A2142G and A2143G mutations, the amplified products were digested with MboII and BsaI restriction enzymes. Figure 1 shows wild type samples digested with MboII and BsaI, respectively: since no recognition site for MboII was present, amplicons appear unmodified (Fig. 1A); moreover, the 768-bp fragment naturally presents a target sequence for BsaI which generates two restriction fragments (108 and 660bp) (Fig. 1B). Strains carrying the A2142G and A2143G mutations are shown in Figure 2. In the presence of the A2142G mutation the resulting restriction DNA fragments were of 418bp and 350bp (Fig. 2A) whereas in the presence of the A2143G mutation the resulting fragments were of 108, 310 and 350bp (Fig. 2B). No differences in restriction patterns were found between both types of samples. Hence, we used the rapid urease test biopsies to study the prevalence of mutations causing clarithromycin resistance in our region. Thirteen out of 62 strains (20.9%) were resistant to clarithromycin: 7/13 (53.8%) harbored the A2142G mutation whereas 6/13 (46.2%) carried the A2143G point mutation. There were no strains harboring a double mutation. In addition, a multiplex PCR amplification for cagA, vacA and babA2 virulence factors was performed on these mutant strains. Figure 3 is a representative image of such assay showing different genotypes of H. pylori. Table 1 shows the distribution of the different virulence factors found in samples carrying point mutations responsible for clarithromycin resistance. It can be observed that vacA m1s1 was the most frequent genotype among the resistant strains (8/13).
Helicobacter pylori clarithromicyn resistance analysis by PCR-RFLP. DNA was purified from biopsies for the rapid urease test (RUT) and for molecular analysis (MA). Amplicons (768bp) from wild type strains were digested with MboII (A) and BsaI (B). M, molecular size marker (Cien Marker, Biodynamics, Buenos Aires, Argentina).
Association between virulence factors and mutation points in the 23S rRNA gene in Helicobacter pylori.
| UPa | cagA | vacA | babA2 | Point mutations | ||
|---|---|---|---|---|---|---|
| s | m | A2142G | A2143G | |||
| UP01 | − | 2 | 2 | − | − | + |
| UP02 | − | 1 | 1 | − | + | − |
| UP03 | − | 2 | 2 | + | − | + |
| UP04 | − | 2 | 1 | − | − | + |
| UP05 | − | 2 | 2 | − | − | + |
| UP06 | + | 1 | 1 | + | + | − |
| UP07 | − | 2 | 1 | + | + | − |
| UP08 | − | 1 | 1 | − | + | − |
| UP09 | − | 1 | 1 | − | − | + |
| UP10 | − | 1 | 1 | + | + | − |
| UP11 | − | 1 | 1 | + | + | − |
| UP12 | − | 1 | 1 | − | − | + |
| UP13 | − | 1 | 1 | + | + | − |
The prevalence of clarithromycin resistance in H. pylori is increasing worldwide and this is the major cause of treatment failure of H. pylori infection. The high consumption of clarithromycin, especially in the treatment of respiratory tract diseases, is probably the main reason of this increased resistance19,23,24,29,30. It is well known that the prevalence of H. pylori resistant to clarithromycin varies among the different countries. The resistance rate in Japan was around 25.9% in 2014; in Europe, the lowest clarithromycin resistance was reported in Norway (5.9%), while the highest in Spain (32.01%) and Portugal (42.35%). Recently, Ghotaslou et al.11 reported the clarithromycin resistance rates in North America (30.8%) and South America (12.9%). Many Latin American countries exhibit a high rate of H. pylori infection and associated diseases with a global clarithromycin resistance prevalence of 13%, with high heterogeneity between different studies7. Since the latest Maastricht Guidelines recommended clarithromycin-containing schemas as first-line therapy in regions with low clarithromycin resistance (<15–20%)16, it is important to know the local susceptibility of H. pylori isolates to guide empirical treatment in clinical practice. Although point mutations in target genes are responsible for antibiotic resistance, in routine clinical microbiology the prevalence of resistance is often determined by phenotypic methods. It is so because new mechanisms of resistance not related to point mutations at antimicrobial target genes can emerge4 and, moreover, the prevalence of known mutations in the target genes may shift to novel ones. Furthermore, even if a positive test result of a genotypic method in one biopsy confirms resistance, it must be considered that a negative result does not rule out resistance, due to the fact that it is possible that isolates in different biopsies evolve in an independent fashion.
Nevertheless, due the slow growing properties of H. pylori, and the specific culture conditions that are needed, the phenotypic approach is challenging and time-consuming. Hence, the usefulness of the molecular approaches to detect point mutations related to clarithromycin-resistance directly from biopsies such as those presented here is very important mainly for low-income clinical laboratories, avoiding the difficulty of phenotypic methods. Moreover, molecular analyses have the advantage of avoiding the effect of heteroresistance in a single niche that can influence MIC results.
Recently, Megraud20 have highlighted the importance of molecular approaches to identify H. pylori antimicrobial resistance, especially for tailored therapies, because when point mutations at 23S-rRNA are detected, other empiric therapies can be employed. Taking this fact into account, continuous surveillance of the prevalent mutations could also be carried out, including their detection by genotypic methods. Such approach has been recently used by Ducournau et al.9 In this regard, PCR-based commercial methods are currently available to rapidly detect resistance to clarithromycin, providing accurate and convenient information. However, they are still too expensive for a low-income setting. In this study we decided to evaluate if the biopsy specimens used for the rapid urease test could be a useful samples to detect resistance to clarithromycin by PCR-RFLP. We found that PCR-RFLP performed from biopsy specimens submitted to the rapid urease test was appropriate to detect resistance to clarithromycin and thus, it was used to analyze clarithromycin resistance in patients infected with H. pylori in our region. In Argentina there are few reports about clarithromycin resistance, however current evidence indicates that such resistance is increasing7,24,29–31. In accordance with those reports, the results of our study showed mutations responsible for clarithromycin resistance in 20.96% of adult patients. However, it must be considered that this percentage could be increased if point mutations other than those studied in our work are taken into account. Unlike other authors1,19,22,26, we did not find a marked predominance between the two mutations, perhaps due to the small number of samples studied. Our results agree with those by Klesiewicz et al.15, who reported the same frequencies of point mutations (A2142G/A2143G) among clarithromycin-resistant H. pylori strains.
Several authors have focused on the relationship among antibiotic resistance and virulence factors of H. pylori with contradictory results. Agudo et al.2, in Spain, found that clarithromycin-resistant H. pylori strains carried the vacA s2/m2 genotype most frequently and were most likely to be cagA-negative than susceptible strains. On the contrary, Rasheed et al.23 found a high proportion of cagA-positive and vacA s1m1 genotypes in H. pylori isolates, thus indicating antibiotic resistance.
A negative association between the prevalence of virulent (cagA+) H. pylori strains and the prevalence of clarithromycin resistance has been reported whereas Boyanova et al.5 did not find an association between point mutations (A2142G and/or A2143G) and the cagA status or the vacA S and M alleles; however a close relationship is observed between A2142G/vacAi1 and A2143G/vacAi2. In regard to the babA2 gene, significant rates of clarithromycin resistance were observed by Karabiber et al.14 in babA2- (and cagE-, iceA1-, and vacA s1c-positive) pediatric patients. On the contrary, Alarcón-Millán et al.3 found no significant differences in genotype distribution and clarithromycin resistance or susceptibility. In addition to this complex scenario, two interesting papers show that the co-existence of diverse genotypes of putative virulence factors in a single host and/or microevolution phenomena must be considered when drawing a correlation with the clinical presentation (and perhaps with resistance patterns)17,18.
In our study, like in other reports, vacA m1s1 was the most frequent genotype among the resistant strains6,29. Due to the fact that – among the vacA genotypes – the vacA m1s1 genotype is the most virulent and that previous reports described higher eradication rates when more virulent strains are present5,23,25,28 this could emphasize the need for fast and reliable ways to identify, at molecular level, the strains present in H. pylori-infected patients in order to select the best eradication treatment and prevent future complications such as pre-neoplastic lesions or progression to gastric cancer. To our knowledge, this is the first study on the analysis of H. pylori clarithromycin resistance (and virulence factor distribution) by molecular methods directly from gastric biopsies in this region of Argentina. In addition, this approach is especially useful in cases where the number or the size of biopsies are limited. Currently, we are devoted to optimizing an allele-specific primer-PCR (ASP-PCR) protocol in order to simplify the detection of clarithromycin resistance in H. pylori.
Conflict of interestThe authors declare that they have no conflicts of interest.
Authors thank Secretaria de Ciencia y Técnica, Universidad Nacional del Litoral (Project PI. 501 201101 00089 LI. 16).