Candida glabrata is a yeast that can cause hazardous fungal infections with high mortality and drug resistance.
AimsThe aim of this study was to determine the profile of drug susceptibility in clinical isolates of C. glabrata and review the resistance mechanisms to caspofungin.
MethodsA total of 50 C. glabrata clinical isolates from Iran were tested for in vitro susceptibilities to amphotericin B, caspofungin, fluconazole and voriconazole. To investigate the mechanism of resistance to caspofungin, hotspot areas of FKS1 and FKS2 genes were sequenced and gene expression profile was evaluated.
ResultsAll the isolates were susceptible to amphotericin B and caspofungin. Fluconazole resistance was exhibited in four isolates. In addition, only one isolate was resistant to voriconazole. FKS2 with 12 point mutations showed more mutations compared to FKS1 that had only two mutations. All substitutions were synonymous. FKS genes were expressed at comparable levels (no statistical significance) in caspofungin-treated and non-treated cultures.
ConclusionsThe silent mutations in the hotspot areas of FKS genes and inconsiderable changes in gene expression were not associated with increased MIC (0.25μg/ml). Other mechanisms of resistance which include mutations outside the hotspot area of FKS genes could be involved in a slight increase of MIC, and they should be identified through complete FKS gene sequencing.
Candida glabrata es una levadura que puede causar infecciones de una alta mortalidad, además de presentar resistencia a los tratamientos antifúngicos.
ObjetivosEl objetivo del presente estudio fue determinar la sensibilidad a diversos antifúngicos de aislamientos clínicos de C. glabrata, además de revisar sus mecanismos de resistencia a la caspofungina.
MétodosSe estudió la sensibilidad in vitro de 50 aislamientos clínicos de C. glabrata de Irán a los antifúngicos anfotericina B, caspofungina, fluconazol y voriconazol. Con el fin de investigar el mecanismo de resistencia a la caspofungina se secuenciaron secciones hotspot de los genes FKS1 y FKS2, y se evaluó su grado de expresión.
ResultadosTodos los aislamientos fueron sensibles a la anfotericina B y a la caspofungina. Cuatro aislamientos fueron resistentes al fluconazol. Un solo aislamiento presentó resistencia al voriconazol. FKS2 mostró un mayor número de mutaciones (12) que FKS1, donde se encontraron solo dos; todas las sustituciones fueron silentes. No se encontró diferencia estadística en la expresión de los genes FKS entre los aislamientos crecidos en cultivo con caspofungina y los que crecieron en cultivo sin la presencia del antifúngico.
ConclusionesLas mutaciones silentes en las areas hotspot de los genes FKS y los cambios insignificantes en la expresión de los genes no se asociaron con un incremento de la MIC (0,25μg/ml). Otros mecanismos de resistencia, como mutaciones fuera de las áreas hotspot de los genes FKS, podrían conllevar un ligero aumento de la MIC, pero para determinar claramente su papel debería realizarse una secuenciación completa de los genes FKS.
Candida species are the most common cause of invasive mycotic diseases in hospitalized patients, and approximately 10% of all nosocomial bloodstream infections worldwide are attributed to this genus.5 Over the past few decades, C. glabrata has emerged as a prominent and potentially multidrug-resistant Candida species,25 such that this organism is the third most common cause of bloodstream candidiasis in Iran.5
Echinocandins are considered to be active drugs against C. glabrata and have been introduced as the first-line treatment of infections associated with C. glabrata,12,15 although several studies have increasingly reported resistance to echinocandins in Candida isolates.6,11,20,23–25 Nevertheless, despite the increasing use of echinocandins, resistance to these drugs is still rare.17,18 Molecular mechanisms of resistance to echinocandins have been previously described.7 In most cases, the molecular mechanisms of resistance are associated with mutations in hotspot areas of FKS genes, and the MIC depends on the position of the mutation and the type of substitution in the translated amino acid.8,13,19,20 The actual prevalence of FKS mutations and echinocandins resistance in each region depends on the local epidemiological and population structure studies. In addition, due to the clinical significance of C. glabrata and the few studies on echinocandins resistance conducted in Iran, this study aimed to determine the profile of drug resistance in clinical isolates of C. glabrata, and review the resistance mechanisms to caspofungin.
Materials and methodsIn this study, 50 clinical isolates of C. glabrata, which had been previously reported, were used.2 Isolates were tested for in vitro susceptibilities to amphotericin B, caspofungin, fluconazole and voriconazole using broth microdilution method as described by the Clinical Laboratory Standards Institute (CLSI) M27-A3 and M27-A4 document guidelines.3,4 Thereafter, isolates that had the highest MIC against caspofungin were used for subsequent analyses. The conventional phenol-chloroform extraction method was used for DNA extraction.10 Then hotspot 1 and hotspot 2 regions of FKS1 and FKS2 genes were amplified and sequenced in both forward and reverse directions with the same primer used for the PCRs.23 Sequencing reactions were performed using BigDye terminator technology (ABI, Foster City, CA) with an ABI Prism 3730 (ABI series) DNA sequencer. Nucleotide sequences were defined by alignment of forward and reverse sequences using MEGA software, version 5.2, and polymorphic sites were confirmed by visual examination of the chromatograms. Then results were compared with DNA database of GenBank (XM-446406 for FKS1 gene and XM-448401 for FKS2 gene).
First, C. glabrata isolates were grown overnight at 35°C in YPD broth medium containing caspofungin, with shaking at 150rpm, and cell cultures without drug treatment served as controls. Total RNA was extracted using RNX-plus kit according to the manufacturer's instruction (SinaClon BioScience Co., Karaj, Iran). Gene expression profile was performed using Two-step SYBR green quantitative reverse transcription-PCR (qRT-PCR) with Applied Biosystems Step One Plus Real-Time PCR System.6 All experiments were performed in triplicate and the evaluation of gene expression was done using Pfaffl method.16 The C. glabrata URA3 gene (GenBank accession no, AY771209) was used for normalization.6
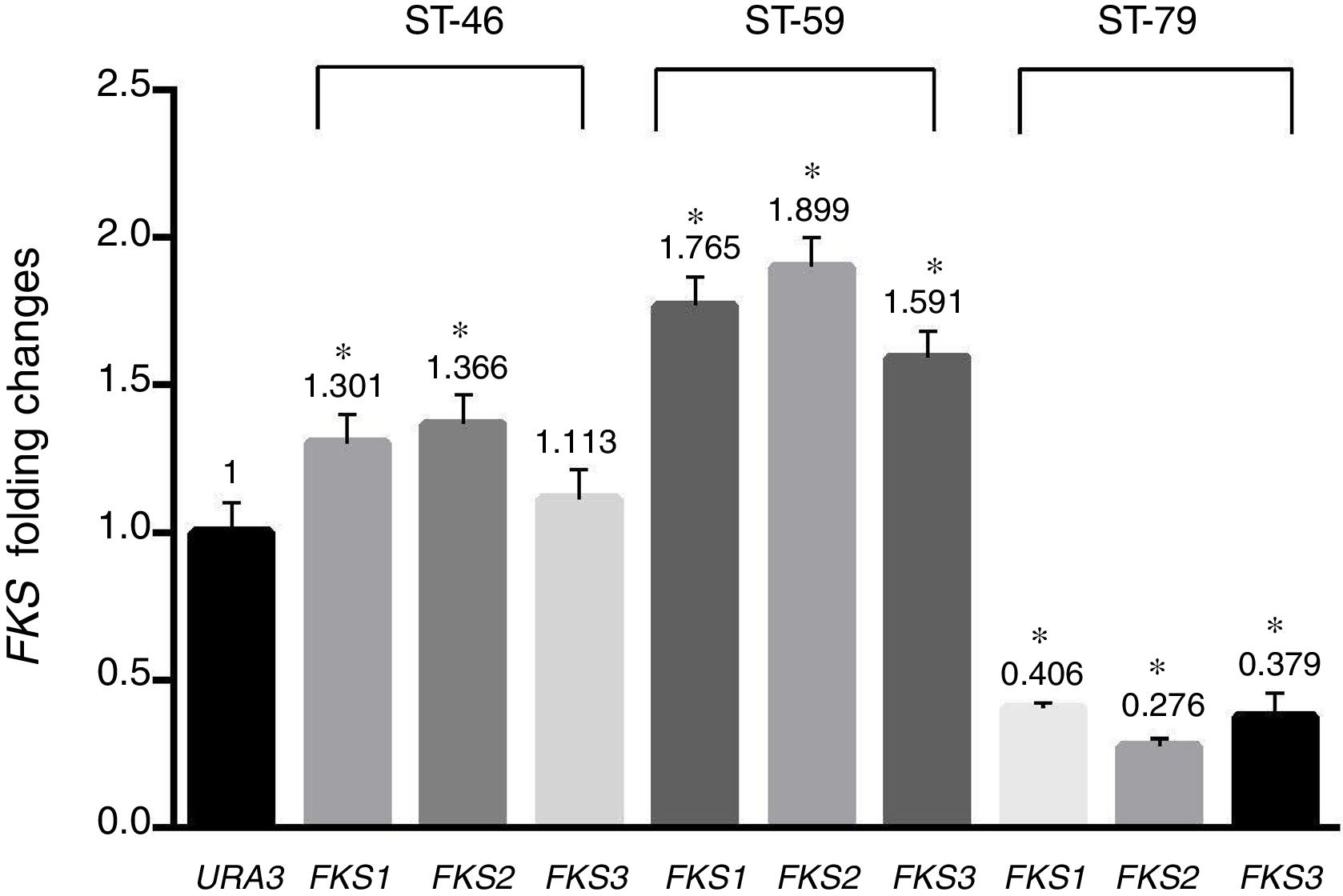
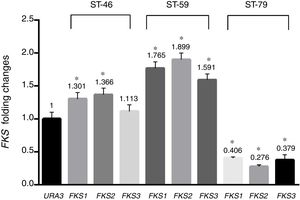
Results and discussionAs shown in Table 1, a total of 41 isolates (82%) were susceptible to fluconazole, five isolates (10%) were susceptible-dose-dependent (MIC=32μg/ml), and four isolates (8%) with MIC=64μg/ml were determined as resistant to fluconazole. In addition, 46 isolates (92%) were susceptible to voriconazole, three isolates (6%) with MIC=2μg/ml were considered susceptible-dose-dependent, and one isolate with MIC=4μg/ml was resistant. All isolates were susceptible to amphotericin B and MIC of all of them was under the epidemiological cutoff (2μg/ml). As shown in Table 1, no isolate resistant to caspofungin was found, but hotspot areas of FKS1 and FKS2 genes of two isolates with MIC of 0.25μg/ml and one isolate with MIC of 0.0625μg/ml were sequenced (GenBank accession numbers MF538706 to MF538717) and compared to the genome database sequences (GenBank accession number XM_446406 for FKS1 and XM_448401 for FKS2). Sequence analysis revealed that all mutations were silent. Nucleotide polymorphic sites and subsequent amino acid substitutions are shown in Table 2. The highest number of polymorphisms occurred in the nucleotide sequence of FKS2 hotspot 1. There were 10 nucleotide replacements and all changes were synonymous. In the hotspot 2 area of FKS2 gene, two variations were also recorded and, finally, in hotspot 1, two sites of FKS1 gene and one nucleotide change were observed. As shown in Fig. 1 the expression level of FKS genes remained relatively stable compared to the URA3, and gene expression in treated and non-treated cultures was not statistically significant.
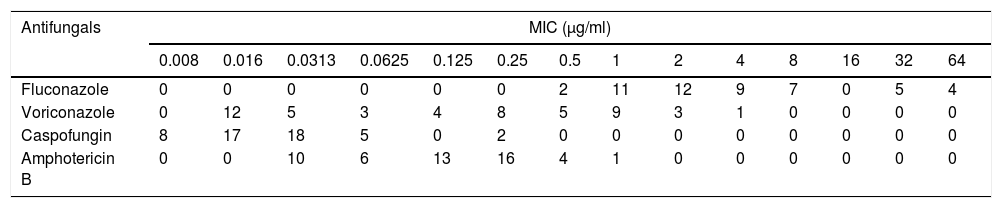
Distribution of MICs for 50 clinical isolates of C. glabrata.
| Antifungals | MIC (μg/ml) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.008 | 0.016 | 0.0313 | 0.0625 | 0.125 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | |
| Fluconazole | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 11 | 12 | 9 | 7 | 0 | 5 | 4 |
| Voriconazole | 0 | 12 | 5 | 3 | 4 | 8 | 5 | 9 | 3 | 1 | 0 | 0 | 0 | 0 |
| Caspofungin | 8 | 17 | 18 | 5 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Amphotericin B | 0 | 0 | 10 | 6 | 13 | 16 | 4 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
Results are presented as the number of C. glabrata isolates with MIC in each two-fold serial dilution of the tested antifungals (concentration: 0.008–64μg/ml).
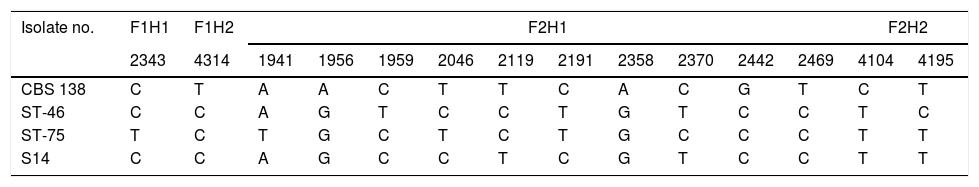
Polymorphic nucleotide sites in hotspot areas of FKS1 and FKS2 compared to the genome database sequences of C. glabrata CBS 138.
| Isolate no. | F1H1 | F1H2 | F2H1 | F2H2 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2343 | 4314 | 1941 | 1956 | 1959 | 2046 | 2119 | 2191 | 2358 | 2370 | 2442 | 2469 | 4104 | 4195 | |
| CBS 138 | C | T | A | A | C | T | T | C | A | C | G | T | C | T |
| ST-46 | C | C | A | G | T | C | C | T | G | T | C | C | T | C |
| ST-75 | T | C | T | G | C | T | C | T | G | C | C | C | T | T |
| S14 | C | C | A | G | C | C | T | C | G | T | C | C | T | T |
F: FKS gene, H: hotspot.
The numbers represent the positions of the variable nucleotides relating to the fragments sequenced.
In this study, no resistance to AMB or caspofungin was found among the studied isolates, and these drugs are recommended in the treatment of C. glabrata infections. All fluconazole-resistant isolates showed low MICs to caspofungin and amphotericin B. On the other hand, three isolates showed high MICs to both fluconazole and voriconazole. Although the sensitivity of C. glabrata to voriconazole is high, an increasing resistance to this drug is possible according to the ECV index and the expanded use of voriconazole in the general population.
There are conflicting results from different studies on echinocandins resistance. Some studies reported that despite the increasing use of echinocandins, resistance to these drugs is still rare.17,18 Other studies demonstrate the emergence of Candida isolates resistant to echinocandins.6,11,20,23–25 Despite the increasing number of reports that have mentioned echinocandins resistance,6,11,20,23–25 this study show that C. glabrata has high sensitivity to echinocandins, and the sensitivity rate to caspofungin was 100%. Considering the low use of echinocandins, such as caspofungin, in Iran such result was hence predictable. The constant exposure to antibiotics can cause the development of drug resistance. In this study, as well as in previous reports, it has also been demonstrated that echinocandins resistance can occur under conditions of exposure to the drug.23 Shields et al. reported that in clinical isolates of C. glabrata, any mutation in FKS genes is associated with prior exposure to echinocandins.22 Alexander et al. also showed that any previous treatment with echinocandins in patients with candidaemia is a significant predictor of FKS mutations in C. glabrata.1
The gene expression level was not statistically different in the treated and non-treated states. FKS mutation rate in C. glabrata has been reported in the range of 3–18%.1,22FKS2 mutations in this species are about two times higher in comparison to FKS1 mutations.9 We found 12 mutations in FKS2 (10 point mutations in hotspot 1, and 2 mutations in hotspot 2) and only 2 mutations in FKS1. All substitutions were synonymous and protein coding was not changed. Therefore, any slight increase in the MIC of the studied isolates would be related to mutations in areas outside the hotspot of FKS genes, and this should be identified through the complete sequencing of FKS gene. Some researchers believe that identifying mutations in FKS genes is the most accurate method for predicting treatment failure with echinocandins.13,19 In most cases, the MIC value in mutant strains was markedly higher than that in wild type strains; a new indicator of clinical interpretive breakpoints distinguishes between different FKS mutations based on their clinical relevance and is also used to predict the response to the treatment.1 Overall, in vitro susceptibility tests are useful in predicting treatment response, therefore it is recommended in high-risk patients. Antifungal susceptibility tests should be routinely conducted,14,21 and echinocandins should be used with caution against Candida species.
Conflict of interestsThe authors declare they have no conflict of interest.
We would like to express our gratitude to the Research Deputy of Tarbiat Modares University for funding our research.