Cryptococcosis is a systemic opportunistic mycosis, caused by Cryptococcus neoformans and Cryptococcus gattii, which affects mainly the central nervous system of immunocompromised patients; no reports have been made on the isolation of the fungus from the environment of Popayán, Colombia.
AimsThe main objective of this investigation was to determinate the incidence of C. neoformans in the urban perimeter in the City of Popayán, Colombia.
MethodsA total of 303 samples from droppings of Columba livia and Bubulcus ibis were collected between September 2012 and June 2013. The samples were processed by conventional techniques; identification of colonies was performed by biochemical tests, and molecular patterns were determined by PCR fingerprinting with the primer (GTG)5 and restriction fragment length polymorphism (RFLP) of the gene URA5.
ResultsA total of 118 (38.94%) samples were positive for Cryptococcus in excreta of C. livia, and 361 strains belonging to Cryptococcus neoformans var. grubii were isolated. From the latter, 99.2% corresponded to the molecular pattern VNI and 0.8% to VNII, with an increased occurrence (24.4%) at a temperature of 22.5°C and a humidity of 60.8%. The excreta of B. ibis did not show the presence of the fungus.
ConclusionsC. livia excreta is a key environmental niche for C. neoformans var. grubii, type VNI, supporting growth and reproduction, and serving as a major source of infection for susceptible populations in Popayán. This represents the first report on the isolation of the agent of cryptococcosis from the environment in this region, with a significant prevalence in bird excreta.
La criptococosis es una micosis sistémica oportunista provocada por Cryptococcus neoformans y Cryptococcus gattii; afecta principalmente al sistema nervioso central de pacientes inmunodeficientes. No existen publicaciones sobre el aislamiento del hongo en la región de Popayán, Colombia.
ObjetivosEl objetivo principal de este estudio fue establecer la incidencia de C. neoformans en el perímetro urbano de la ciudad de Popayán, Colombia.
MétodosSe recogió un total de 303 muestras de materia fecal de las aves Columba livia y Bubulcus ibis entre septiembre de 2012 y junio de 2013. Las muestras se procesaron con técnicas convencionales de cultivo; la identificación de las colonias se realizó mediante pruebas bioquímicas, y los patrones moleculares se establecieron mediante PCR con el cebador (GTG)5 y la técnica de polimorfismos de longitud de los fragmentos de restricción (RFLP, por su abreviatura en inglés) del gen URA5.
ResultadosUn total de 118 (38,94%) muestras de excrementos de C. livia fueron positivas para Cryptococcus y se recuperaron 361 aislamientos pertenecientes a Cryptococcus neoformans var. grubii, de los cuales el 99,2% correspondían al patrón molecular VNI y el 0,8% al VNII, con un aumento en la incidencia (24,4%) a una temperatura de 22,5°C y una humedad del 60,8%. No hubo crecimiento de este hongo en los excrementos de B. ibis.
ConclusionesLos excrementos de C. livia son un nicho ambiental fundamental para C. neoformans var. grubii, tipo VNI; ello permite su crecimiento y reproducción, y que sean una potencial fuente de infección para la población susceptible de Popayán. Esta es la primera publicación sobre el aislamiento del agente etiológico de la criptococosis en el entorno de esta región, con una prevalencia considerable en los excrementos de aves.
Cryptococcosis, caused by the encapsulated yeasts Cryptococcus neoformans and Cryptococcus gattii, is an opportunistic systemic disease of global distribution that mainly affects patients with medical conditions in their immune systems. The clinical presentation may vary from asymptomatic pulmonary colonization to disseminated disease affecting multiple organ systems, spreading and infecting any part of the body, with meningoencephalitis being the leading cause of death by this infection.42
C. neoformans comprises C. neoformans var. grubii (serotype A), C. neoformans var. neoformans (serotype D), and a hybrid serotype AD; C. gattii includes serotypes B and C. Recently, hybrids between serotypes B and D, and A and B have been reported.8,25 According to several studies, these two species have epidemiological, ecological, molecular, geographic and clinical characteristics different enough to be considered distinct.21,40,41 Recently, a new classification was proposed, based on genotypic and phylogenic analyses of 11 loci, suggesting that the current C. neoformans var. grubii and C. neoformans var. neoformans are two separate species, and five species existing within the designation of C. gattii.30
In Colombia, slight epidemiological information exists concerning cryptococcosis. Nevertheless, several environmental studies have been carried out in different cities of Colombia; in 1994, Cryptococcus was found in 53.8% of excreta samples obtained in different cities in Colombia.23 Two years later, a study carried out on excreta of pigeons in the urban perimeter of the city of Cali indicated the presence of C. neoformans, with C. neoformans var. neoformans present in 49.6% of the cases, demonstrating that this yeast has a wide distribution in the city.5
In 2005 a study was carried out in the Department of Cundinamarca, in an attempt to determine the relationship between the environmental distribution of the different varieties of C. neoformans and the thermal floors of the Department of Cundinamarca. Of a total of 765 samples from 26 municipalities, 104 samples were positive for C. neoformans with a frequency of 31% for serotype A, 59% for serotype B and 10% for serotype C.45
Birds are one of the main transmitters of several diseases, acting as hosts of pathogenic microorganisms and playing a vital role in the dissemination of the etiologic agents. Birds’ excreta can contaminate the air,4 which contributes to the distribution and transmission of infectious propagules.9
Different investigations indicate that the excreta of pigeons (Columba livia) are the main source for C. neoformans infections in urban areas, but have also been associated with pathogens that cause aspergillosis, salmonellosis, listeriosis and staphylococcosis.27C. neoformans has been mainly isolated from sites with accumulations of pigeon droppings, such as cornices, domes, eaves of buildings and attics of old buildings. These habitats have characteristics such as moisture, alkalinity and nitrogen substances, allowing the yeast to remain viable for more than two years.49 The common pigeon (C. livia), like many birds, has a high body temperature that allows the microorganism to survive, but not to develop.
The purpose of this research focuses on the determination of the distribution of C. neoformans in the environment of the urban perimeter of the City of Popayán, Cauca, Colombia.
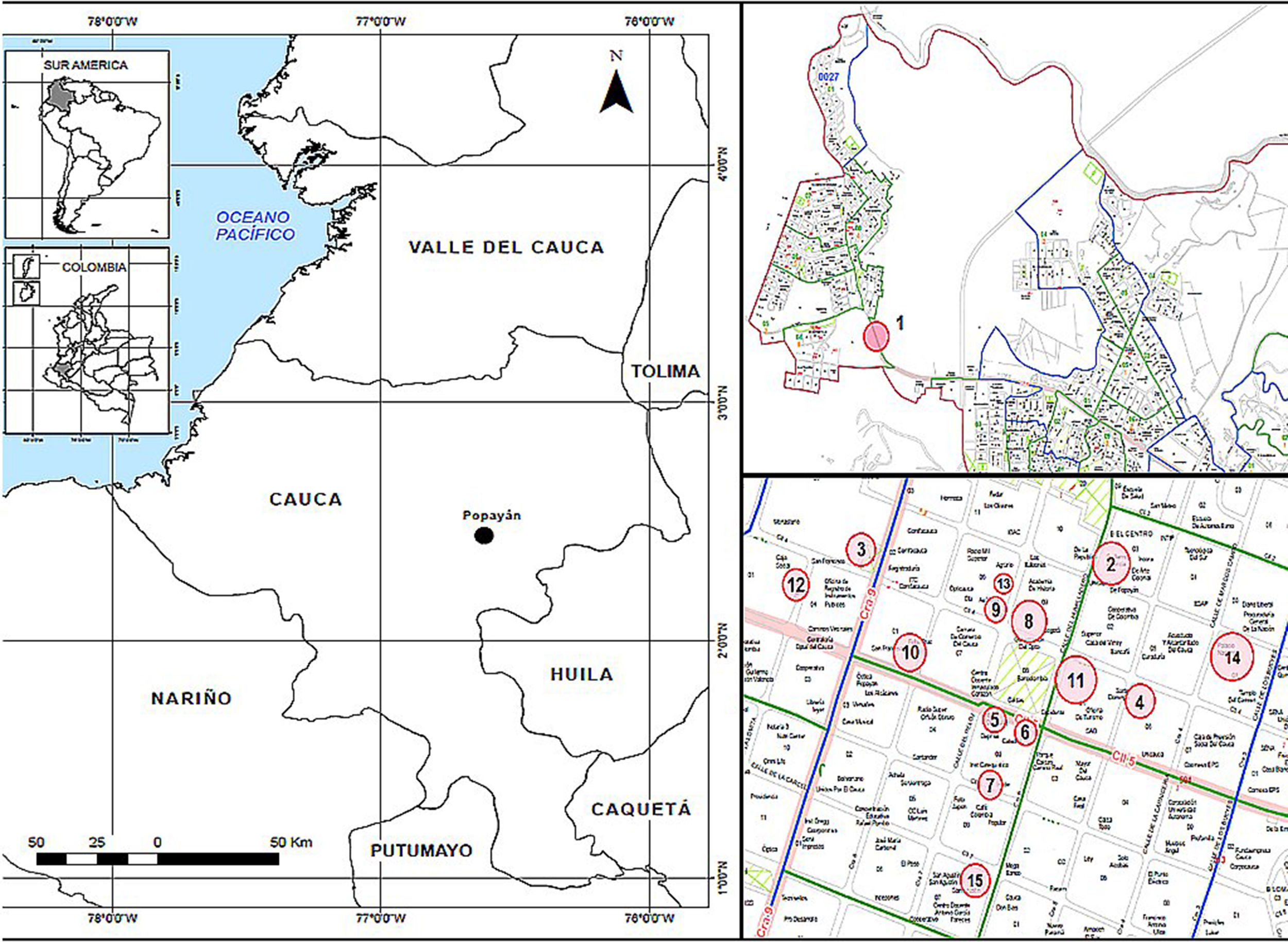
Materials and methodsStudy areaThe City of Popayán, capital of the department of Cauca, is located in the Valley of Pubenza, between the Western and Central mountain ranges. Its coordinates are N 2°26′39″ O 76°37′17″, and its altitude is 1737meters above sea level (MASL). Because it is very close to the equatorial line, it has an average temperature of 18–19°C throughout the year, with peak temperatures in the months of July to September (high temperatures of 29°C and minimum of 10°C). It has 265,702 inhabitants, according to the census reported by the National Statistics Administrative Department (DANE) taken in 2005.17 The city spans 512km2, and it has an annual rainfall of 1941mm. It also has extensive flat and undulating areas, mainly located near the Cauca River. The municipality is predominantly urban; 90% of the population occupies this area and the remaining 10% occupies the rural area. It is divided into 9 districts, 23 townships.
Collection and processing of samplesThere were found different reference points, such as roofs, cupolas, dovecotes, squares and parks, with a high population of the species Bubulcus ibis (cattle egret) and C. livia (Common pigeon). The sampling was performed by dividing the urban perimeter into 9 sectors, and inspecting exhaustively those sites with a high concurrence of people and a high population of B. ibis and C. livia (over 40 specimens per site).
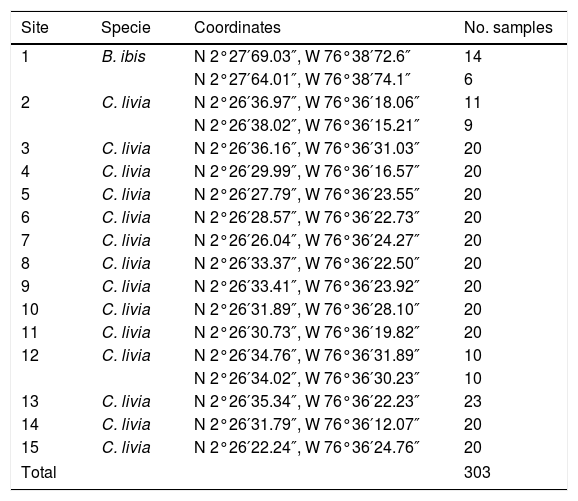
Two sectors were chosen, which were divided into 15 sites (Table 1 and Fig. 1). Two hundred eighty three C. livia excrement samples and 20 B. ibis excrement samples (303 samples) were collected at these sites. From each sample, 15g of excreta were collected in plastic bags. Additionally, environmental factors were recorded at each sampling site (Table 1), including altitude and geographical position, taken with a GPS Garmin Etrex 30; the temperature and relative humidity were taken with a precision thermo-hygrometer ANVI.
Sampling sites for the isolation of Cryptococcus neoformans (coordinates are shown), the bird species at each site, and the number of samples collected; sites 1, 2 and 12 were sampled twice in different days to complete the total of representative samples.
| Site | Specie | Coordinates | No. samples |
|---|---|---|---|
| 1 | B. ibis | N 2°27′69.03″, W 76°38′72.6″ | 14 |
| N 2°27′64.01″, W 76°38′74.1″ | 6 | ||
| 2 | C. livia | N 2°26′36.97″, W 76°36′18.06″ | 11 |
| N 2°26′38.02″, W 76°36′15.21″ | 9 | ||
| 3 | C. livia | N 2°26′36.16″, W 76°36′31.03″ | 20 |
| 4 | C. livia | N 2°26′29.99″, W 76°36′16.57″ | 20 |
| 5 | C. livia | N 2°26′27.79″, W 76°36′23.55″ | 20 |
| 6 | C. livia | N 2°26′28.57″, W 76°36′22.73″ | 20 |
| 7 | C. livia | N 2°26′26.04″, W 76°36′24.27″ | 20 |
| 8 | C. livia | N 2°26′33.37″, W 76°36′22.50″ | 20 |
| 9 | C. livia | N 2°26′33.41″, W 76°36′23.92″ | 20 |
| 10 | C. livia | N 2°26′31.89″, W 76°36′28.10″ | 20 |
| 11 | C. livia | N 2°26′30.73″, W 76°36′19.82″ | 20 |
| 12 | C. livia | N 2°26′34.76″, W 76°36′31.89″ | 10 |
| N 2°26′34.02″, W 76°36′30.23″ | 10 | ||
| 13 | C. livia | N 2°26′35.34″, W 76°36′22.23″ | 23 |
| 14 | C. livia | N 2°26′31.79″, W 76°36′12.07″ | 20 |
| 15 | C. livia | N 2°26′22.24″, W 76°36′24.76″ | 20 |
| Total | 303 | ||
Most of the samples were taken from nests and feeding sites of these species. The excreta of birds were characterized by colors between green and white/yellowish when it was recent, and between reddish and brown after some time.
Subsequently, 5g of each sample previously collected were put in a 50ml Falcon tube, and 25ml of sterile PBS (pH 7.4) were added to homogenize the sample. Afterwards, the sample was poured off for 30minutes, and the content of the tube was filtered through a sterile gauze into a new Falcon tube of 15ml. Fifty μl of an antibiotic solution (composed by penicillin, 20mg/l, and streptomycin, 40mg/l) were added to each of the filtrates. All samples were subsequently streaked onto Guizotia abyssinica agar plates with an aliquot of 100μl, and incubated at 28°C. The cultures remained under observation for 30 days.16
IdentificationBrown colonies (phenoloxidase production) were recovered as possible isolates of C. neoformans. The cultures which showed no colony growth were incubated for 30 days, being monitored every 72h. Randomly, when possible, up to 10 colonies were recovered on Sabouraud plates to assess Cryptococcus identity by using microscopic observation and biochemical tests: urease production, inhibition of cycloheximide (sown in Agar Mycosel) and presence of nitrates. Once the genus Cryptococcus was confirmed, species determination was done in canavanine glycine bromothymol blue (CBG) agar.22,48
Molecular testsDNA extraction was performed using the technique described by Casali et al.13 Eight control strains were included in the analysis, as described by Meyer et al.: VNI-VNIV for C. neoformans and VGI-VGIV for C. gattii.39 Molecular patterns of environmental strains were determined by using restriction fragment length polymorphisms (RFLP) of the URA5 gene and PCR fingerprinting with the primer (GTG)5.20 The pattern of each isolate in RFLP was determined according to the protocol described previously. RFLP patterns were visually assigned by comparison with the standard strains (VNI-VNIV and VGI-VGIV).20,40 PCR fingerprinting was carried out according to the procedure described by Meyer et al. Molecular types VNI-VNIV and VGIV-VGI were assigned according to the major bands observed. All bands that were visible were included in the analysis, regardless of their intensity.20,40
StatisticsA database in SPSS version 15 was created: the isolates found and the climatic variables (temperature, humidity and altitude) were included. Contingency tables were constructed to observe trends and proportions. Data related to the environmental variables were analyzed using a one factor ANOVA test, followed by a post hoc analysis using the Tukey contrast hypothesis test. Besides, the results of the environmental variables were grouped into three categories (high, medium and low) and their differences were statistically analyzed using a Pearson chi-squared based contrast hypothesis test.
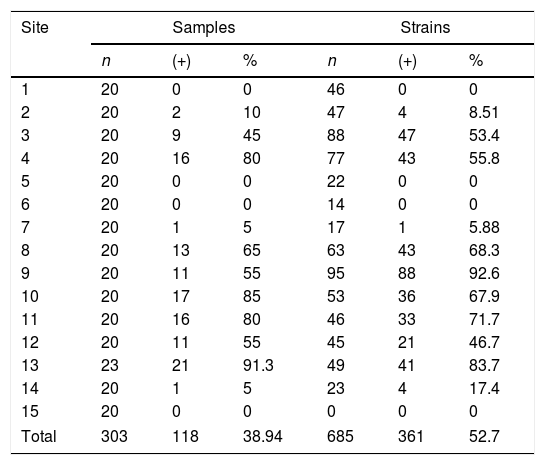
ResultsA total of 303 samples were analyzed, of which 118 (38.9%) samples were positive for C. neoformans, isolating 361 colonies. The samples were collected at 15 sites, and a recovery of 38.9% was observed (Table 2). Sites 1, 5, 6 and 15 with C. livia samples showed no growth of the fungus. In site 1, B. ibis samples were collected, but did not present fungus growth.
Percentage of isolation of Cryptococcus neoformans in relation to the processed samples and strains recovered for each site. The data are grouped by samples (samples of excreta collected per site) and strains (number of colonies analyzed).
| Site | Samples | Strains | ||||
|---|---|---|---|---|---|---|
| n | (+) | % | n | (+) | % | |
| 1 | 20 | 0 | 0 | 46 | 0 | 0 |
| 2 | 20 | 2 | 10 | 47 | 4 | 8.51 |
| 3 | 20 | 9 | 45 | 88 | 47 | 53.4 |
| 4 | 20 | 16 | 80 | 77 | 43 | 55.8 |
| 5 | 20 | 0 | 0 | 22 | 0 | 0 |
| 6 | 20 | 0 | 0 | 14 | 0 | 0 |
| 7 | 20 | 1 | 5 | 17 | 1 | 5.88 |
| 8 | 20 | 13 | 65 | 63 | 43 | 68.3 |
| 9 | 20 | 11 | 55 | 95 | 88 | 92.6 |
| 10 | 20 | 17 | 85 | 53 | 36 | 67.9 |
| 11 | 20 | 16 | 80 | 46 | 33 | 71.7 |
| 12 | 20 | 11 | 55 | 45 | 21 | 46.7 |
| 13 | 23 | 21 | 91.3 | 49 | 41 | 83.7 |
| 14 | 20 | 1 | 5 | 23 | 4 | 17.4 |
| 15 | 20 | 0 | 0 | 0 | 0 | 0 |
| Total | 303 | 118 | 38.94 | 685 | 361 | 52.7 |
The occurrence of C. neoformans varies according to environmental factors. Fungus recovery occurred in a temperature range between 19.8°C and 27.4°C, with an increased occurrence at 22.5°C, while at temperatures of 24.1°C and 25.1°C, there was no growth. Humidity values were between 36% and 76.4%, with the highest occurrence at 60.8%, with some variations in the values of 37.5% and 38.3% in which no C. neoformans growth was observed (Fig. 3). On the other hand, the altitude presented ranges between 1745 and 1757 MASL, where the greatest occurrence of isolation of the yeast was at 1752 MASL.
The temperature presented a statistically significant similarity between Low and Medium categories (p=0.968); there was a greater influence of temperature on the High category. In humidity, a significant difference was observed between the High and Low categories (p=0.012), but not between High and Medium (p=0.658), or Medium and Low (p=0.811). The variable altitude revealed that there is very little difference between the High–Medium (p=0.0012) and Low category (p=0.0011). Sites 1, 2, 5, 6, 7, 14 and 15 were exposed to the sun most of the time (Fig. 2; Table 2). The sites that experienced the most consistent sunlight had the least fungal growth.
Statistical tests demonstrated that the variables of temperature and humidity have a marked influence on the isolation of C. neoformans. Temperature is more influential in the highest category of occurrence (p=0.0012), while humidity is influential in all categories, being grouped into a single subset (by Dunnet T3 analysis). Altitude did not show a statistically significant influence in the recovery of the fungus.
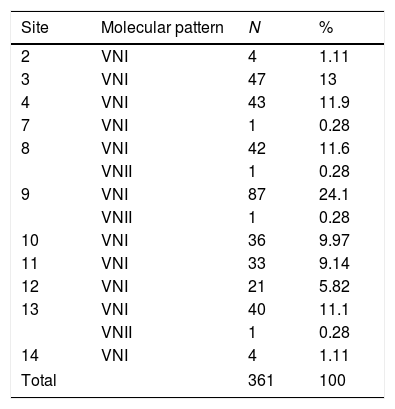
Regarding the biochemical identification, all isolates belonged to C. neoformans. Molecular typing revealed that all isolates were grouped as serotype A; 99.2% (358 isolates) belonged to molecular pattern VNI, and 0.8% (3 isolates) to molecular type VNII (Figs. 3 and 4; Table 3).
Molecular patterns of Cryptococcus neoformans in each sampled site. Locations 1, 5, 6 and 15 do not appear in the table since they showed no growth of C. neoformans.
| Site | Molecular pattern | N | % |
|---|---|---|---|
| 2 | VNI | 4 | 1.11 |
| 3 | VNI | 47 | 13 |
| 4 | VNI | 43 | 11.9 |
| 7 | VNI | 1 | 0.28 |
| 8 | VNI | 42 | 11.6 |
| VNII | 1 | 0.28 | |
| 9 | VNI | 87 | 24.1 |
| VNII | 1 | 0.28 | |
| 10 | VNI | 36 | 9.97 |
| 11 | VNI | 33 | 9.14 |
| 12 | VNI | 21 | 5.82 |
| 13 | VNI | 40 | 11.1 |
| VNII | 1 | 0.28 | |
| 14 | VNI | 4 | 1.11 |
| Total | 361 | 100 | |
This is the first epidemiological investigation that examines both descriptive and molecular data on C. neoformans var. grubii isolated from excreta of C. livia, in the urban perimeter of the city of Popayán, Cauca, contributing to the knowledge about the epidemiology of this fungus in Colombia.
There have been several studies to determine the epidemiology of C. neoformans. This fungus is present in excreta of several species of birds; Emmons published in 1951 the first report connecting C. livia excreta with C. neoformans, and more recently published data on environmental studies show the prevailing presence of this fungus in C. livia excreta.10,15,31,37,47 Recent studies have mentioned the excreta of other bird species as natural habitats of C. neoformans: Falco tinnunculus, Buteo buteo, or species of the families Psittacidae, Cacatuidae and Psittacula.5,14,36,46
As observed in the results obtained in this study, C. neoformans presented an occurrence of 73.3% amongst the sites, and 38.9% amongst all the samples, recovering 361 colonies (49.0%) (Fig. 2; Table 2). It provides a strong support about the presence of this fungus in the City of Popayán. These results are aligned with the data shown in different epidemiological studies made in the region of the Americas. In the United States, El Salvador, Chile and Brazil, an average between 27% and 39% of positive samples were found.3,16,27,33,37,51 The presence of this fungus in the City of Popayán is added to the results obtained in several studies carried out in Colombia, where the presence of C. neoformans has been reported in clinical and environmental samples.3,5,11,20,28 It can also be observed that C. neoformans was found in greater frequency in sites 3, 4, 8, 9 and 13, probably because these sites presented a higher concentration of excreta, given that pigeons built their nests on these sites. It must be noted that C. livia does not remove its excreta from the nest, allowing large stacks of substrate to accumulate, creating optimal conditions for fungal growth, and increasing the density of microorganisms in these sites (Fig. 1; Table 2).
Studies have shown that ultraviolet light inhibits yeast growth,29,35 which likely occurred here as well. In contrast, other studies suggest that the melanin produced by this fungus allows it to survive in sunlight, and that the radiation can be turned into metabolic energy.51 In this study solar radiation seemed to inhibit C. neoformans survival. The microhabitat in each collecting site may also influence the recovery of the fungus. A high exposure to sunlight produces greater desiccation of the microenvironment, as well as an increase of its temperature, altering the ideal requirements for the growth of the fungus.
The concentration of the nutrients present in some sites and excreta could have influenced the recovery of the fungus. Excreta of cattle egret (B. ibis) in site 1 (Figs. 1 and 2) vary in their concentrations of nutrients, so C. neoformans inhibitors44 may be present. The process of digestion in the digestive system of this species must also be considered.47 Furthermore, it is possible that a mix of excreta of B. ibis with the feces of other species that also inhabit this niche (Coragyps atratus; Order Accipitriformes, and some species of mammals in the Chiroptera and Rodentia orders), could have altered nutrients concentrations. The presence of other organisms (molds, bacteria) that grew together with C. neoformans suggest the existence of a competition for the essential niche, being one of the possible factors that altered the population density of C. neoformans as indicated by several investigations.7,9,15,34
The environmental variables showed that the recovery of the fungus is influenced by temperature and humidity. When categorizing the strains recovered there is a significant difference between High and Low categories (Fig. 2). This indicates the important role of conditions necessary for an optimal growth. It is very important to emphasize that this study was developed in the same thermal floor (humid mountainous forest).32 Therefore the variation in altitude among the different places is minimum, in contrast to temperature and humidity, which varied according to environmental and structural conditions of each site (Fig. 2).
Identification of C. neoformans var. grubii strains showed the presence of serotype A for all the isolates and the molecular patterns VNI and VNII (Table 3, Figs. 3 and 4). These results coincide with biochemical analyzes and demonstrates that the same species with two molecular patterns may share the same niche.7,12,18,19,43 The molecular method used does not have a discriminatory power as high as that shown with Multi Locus Sequence Typing (MLST) (9 clades) or Amplified Fragment Length Polymorphism (AFLP) (13 genotypes); therefore, it may be suitable to apply these techniques in the future to further characterize the isolates recovered in this study. According to the literature, an association between AFLP1-AFLP1A may be established with molecular types VNI and VNII, respectively.30 The geographic position of Colombia offers optimal conditions for the setting-up of C. gattii species, which is restricted to tropical and subtropical zones. For example, in the City of Cücuta, Colombia, 68 isolates of C. gattii serotype C were recovered from almond tree detritus12; this species has also been isolated in the Department of Cundinamarca, Colombia.19,28,45 There are evidences that C. gattii is present in the excreta of some birds,1,26,50 which are the fundamental niche of C. neoformans serotype A and D. This demonstrates that the two species can grow together, although the species C. gattii does not have enzymes to counteract the effect of metabolizing creatinine, which also alkalizes the substrate.50 It is also important to consider the relative frequency with which Cryptococcus species produces inter and intra hybrid species, as it has been observed in different studies in patients with infections by hybrids of Cryptococcus.2,6,24,30 The species C. gattii and hybrids of Cryptococcus were not found in the sampled sites in this study, but a more comprehensive and more discriminatory molecular procedure could demonstrate the existence of other species and varieties of this fungus in the city.
These observations are a reflection of studies performed worldwide; excreta of pigeons are one of the main niches of this fungus, responsible for most cases of cryptococcosis associated with AIDS patients in South America.38,50 It is very important to note that the chosen sites, in addition to having a high population of birds in the surrounding areas, have also a high human population (except for site one, which is in the city limits [Fig. 1]) who might be infected by the spread propagules.
This study confirms the presence of C. neoformans var. grubii in excreta of C. livia, which has the ideal conditions for the growth, reproduction and spreading of the fungus, becoming a significant source of Cryptococcus infection. Furthermore, the results show for the first time the presence of C. neoformans in the City of Popayán, which is of great epidemiological importance.
Conflict of interestThe authors declare no conflict of interest in this publication.
The authors would like to thank Departamento Administrativo de Ciencia, Tecnología e Innovación, Colciencias for its financial support (grant 20113600115683), Julieta Montero Carvajal, Coordinator of clinical laboratory specialist at the University of Cauca; and María Fernanda Mesias, laboratory assistant Microbiology and Parasitology, Faculty of Health Sciences, University of Cauca. The authors thank Norida Velez for her support in the statistical analysis.