Matrix-assisted laser desorption-time of flight mass spectrometry (MALDI-TOF-MS) represents a revolution in the identification of microorganisms of clinical interest. Many studies have confirmed the accuracy and fastness of this tool with routine strains.
AimsTo identify clinical isolates of Candida from patients diagnosed with candidemia.
MethodsVitek-MS™ system was used with a collection of 298 blood isolates of the genus Candida represented by 9 different species. Sequencing of the internal transcribed spacer (ITS) region of ribosomal DNA cluster was used as the reference method.
ResultsThe results of Vitek-MS™ were concordant with those obtained with the reference method for 279 (93.62%) isolates (Kappa coefficient (κ)=0.91). Vitek-MS™ misidentified 10 (3.36%) isolates and did not identify 9 (3.02%) isolates.
ConclusionsThis study determines the potential of Vitek-MS™ in yeast identification, being a reliable and fast alternative in the clinical laboratory, with an acceptable sensitivity of 82% (IC 95%: 70–90.6%), in comparison with a 100% (IC 95%: 92.9–100%) sensitivity of the conventional methods.
La espectrometría de absorción de masas mediante láser asistido por una matriz (MALDI-TOF MS) representa una revolución en la identificación de microorganismos de interés clínico. Muchos estudios han confirmado la exactitud y rapidez de esta herramienta con aislamientos de la rutina clínica diaria.
ObjetivosIdentificar aislamientos clínicos del género Candida procedentes de pacientes con un diagnóstico de candidemia.
MétodosSe utilizó el sistema VITEK® MS con un grupo de 298 aislamientos sanguíneos del género Candida, representado por 9 especies diferentes. Se utilizó como método de referencia la secuenciación de la región del espaciador de transcripción interno (ITS, por sus siglas en inglés) del ADN ribosómico.
ResultadosLos resultados de VITEK® MS coincidieron con aquellos obtenidos por el método de referencia en 279 (93,62%) de los aislamientos (coeficiente Kappa [κ]=0,91), mientras que clasificó erróneamente a 10 (3,36%) aislamientos y no identificó otros 9 (3,02%).
ConclusionesVITEK® MS es una alternativa fiable y rápida en la identificación de levaduras en el laboratorio clínico, con una sensibilidad aceptable del 82% (IC 95%:70-90,6%) en comparación con una sensibilidad del 100% (IC 95%:92,9-100%) de los métodos convencionales.
Rapid identification of yeasts is clinically important when treating immunocompromised and hospitalized patients with disseminated yeast infections.10Candida species are opportunistic pathogens causing systemic infections contributing to high mortality in hematological, transplanted and intensive care unit (ICU) patients. Further, some of these species are intrinsically resistant to several antifungal drugs. Accordingly, rapid identification of Candida species would be beneficial for a better management of invasive infections caused by these organisms.11,17,23
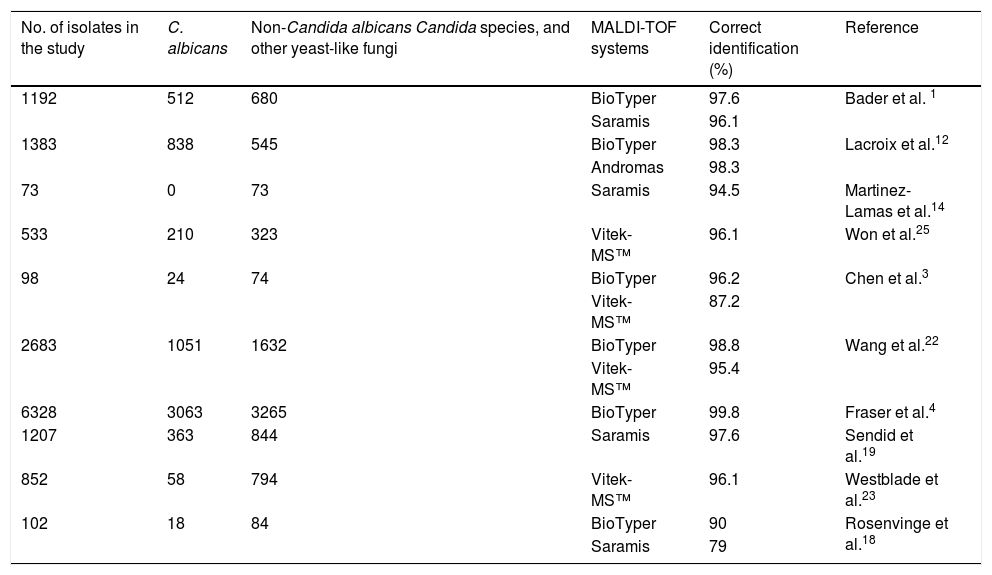
Traditionally, morphological, molecular, biochemical and/or immunological methods have been used for the identification of yeasts. Unfortunately, in most cases, these procedures are laborious and slow.16 Matrix-assisted laser desorption/isolation-time of flight mass spectrometry (MALDI-TOF-MS) is increasingly used in clinical laboratories for fast identification of microorganisms.1,21,25,28 This method is based on a pulsed laser that ionizes particles previously implanted in a matrix. These particles, which travel through the mass analyzer, reach a detector and creates a mass spectrum that is compared to reference spectra in a well-characterized library.3,24 Two MALDI-TOF-MS devices are currently used in most clinical laboratories: BioTyper (Bruker Daltonics, Bremen, Germany) and Vitek-MS™ (bioMérieux, Marcy l’Etoile, France). The performance of both systems for identifying Candida species have been evaluated in previous studies, with correct identification percentage ranges from 90% to 99.8% and 87.2% to 96.1%, for BioTyper and Vitek-MS™, respectively3,4,15,24,27 (Table 1).
Reports on Candida identification by MALDI-TOF systems.
| No. of isolates in the study | C. albicans | Non-Candida albicans Candida species, and other yeast-like fungi | MALDI-TOF systems | Correct identification (%) | Reference |
|---|---|---|---|---|---|
| 1192 | 512 | 680 | BioTyper | 97.6 | Bader et al. 1 |
| Saramis | 96.1 | ||||
| 1383 | 838 | 545 | BioTyper | 98.3 | Lacroix et al.12 |
| Andromas | 98.3 | ||||
| 73 | 0 | 73 | Saramis | 94.5 | Martinez-Lamas et al.14 |
| 533 | 210 | 323 | Vitek-MS™ | 96.1 | Won et al.25 |
| 98 | 24 | 74 | BioTyper | 96.2 | Chen et al.3 |
| Vitek-MS™ | 87.2 | ||||
| 2683 | 1051 | 1632 | BioTyper | 98.8 | Wang et al.22 |
| Vitek-MS™ | 95.4 | ||||
| 6328 | 3063 | 3265 | BioTyper | 99.8 | Fraser et al.4 |
| 1207 | 363 | 844 | Saramis | 97.6 | Sendid et al.19 |
| 852 | 58 | 794 | Vitek-MS™ | 96.1 | Westblade et al.23 |
| 102 | 18 | 84 | BioTyper | 90 | Rosenvinge et al.18 |
| Saramis | 79 |
Our hypothesis is that Vitek-MS™ system can replace the conventional methods for the identification of Candida species with full confidence in a laboratory of clinical microbiology. The aim of the study was to evaluate the performance of this system to identify Candida isolates recovered from bloodstream infections, using molecular methods as the gold standard, especially the ITS sequencing.
Material and methodsStrains and identification by conventional methodsThe present study was conducted at an 800-bed teaching hospital (University Hospital Marqués de Valdecilla, Santander, Spain) which has both an hematological and a transplantation program. We included 298 Candida isolates from blood cultures obtained between January 1st, 2005 and December 31st, 2011. This is a retrospective study on a collection of strains from blood cultures of all hospital services with very different patients, from neonates to elderly people, with an average age of 61.35 years. One isolate per patient was included except when a new episode of candidemia occurred after one month of the previous one. Blood cultures were processed using the Bactec 9240 system (Becton Dickinson, Franklin Lakes, USA).
Candida albicans isolates were preliminarily identified by their growth in both Saboureaud agar with chloramphenicol and Saboureaud agar with chloramphenicol-actidione (Bio-Rad, Hercules, CA, USA), and CHROMagar Candida (Becton Dickinson, Heidelberg, Germany). A molecular method with specific primers was also used.14 Additionally, for every Candida isolate different from C. albicans, the API-ID32C identification system (bioMérieux) was used according to the manufacturer's instructions.6 API-ID32C galleries were incubated at 30°C and results were read after 24h, extending incubation until 48h when recommended by the apiweb™ profile. Ten Cryptococcus neoformans and 40 Rhodotorula mucilaginosa, from a collection of strains, were used as negative controls.
Molecular identificationFor the amplification and sequencing of the ITS (internal transcribed spacer) region of the ribosomal DNA, DNA was extracted from individual colonies grown on CHROMagar plates using the Instagene Matrix (Bio-Rad). The ITS region was amplified with primers ITS1 and ITS4, described previously by White el al.26 The PCR product was purified with NucleoSpin®Gel (Macherey-Nagel, Duren, Germany) and sequenced with ABI PRISM® 377 (Applied Biosystems, Foster City, USA). The sequences of the isolates were compared with those deposited in the GenBank database using BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi). To establish the correct identification, sequences of a given pairwise alignment with the lowest E-value and the highest number of identities (expressed as a percentage) were selected as the most likely species. For C. albicans isolates, instead of sequencing, a molecular method with specific primers14 was used.
In order to determine the concordance between the two identification techniques (one conventional and the other based on a MALDI-TOF-MS system), Kappa coefficient (κ), corresponding to the proportion of concordances observed among the total number of observations, having excluded the concordance produced by random, was calculated.13 Diagnostic parameters results were obtained with Stata®v.14 program.
MALDI-TOF MS™Identification of the organisms with the Vitek-MS™ was performed following the manufacturer's recommendations: cells from a single colony on a CHROMagar Candida plate, incubated for 24h at 37°C, were directly applied onto the steel carrier, lysed by suspension in 0.6μl of 28.9% formic acid (VITEK MS-FA) and dried for a short time (approximately 2min). The sample was allowed to air-dry at room temperature, overlaid with 1μl of α-cyano-4-hydroxycinnamic) VITEK® MS-CHCA matrix and again allowed to air-dry. Measurement was performed on a Vitek-MS™ with SARAMIS MS-IVD v2 database (Anagnos Tee GMBH, bioMérieux), in positive linear mode, with a mass range of 2–20kDa, using Escherichia coli strain ATCC® 8739™ as a molecular mass standard. The intensity of the 50-Hz nitrogen laser was under the control of the acquisition software at the settings recommended by the manufacturer. Two samples of the same strain were applied and only hits within the spectra database with scores of 99.9% and a single shot, without repetitions, were accepted. Results were categorized as “no read” if a bad acquisition (P201error) was obtained in both samples and as “no identification” when the result of the spectra corresponded to a P150 error (not in the database).
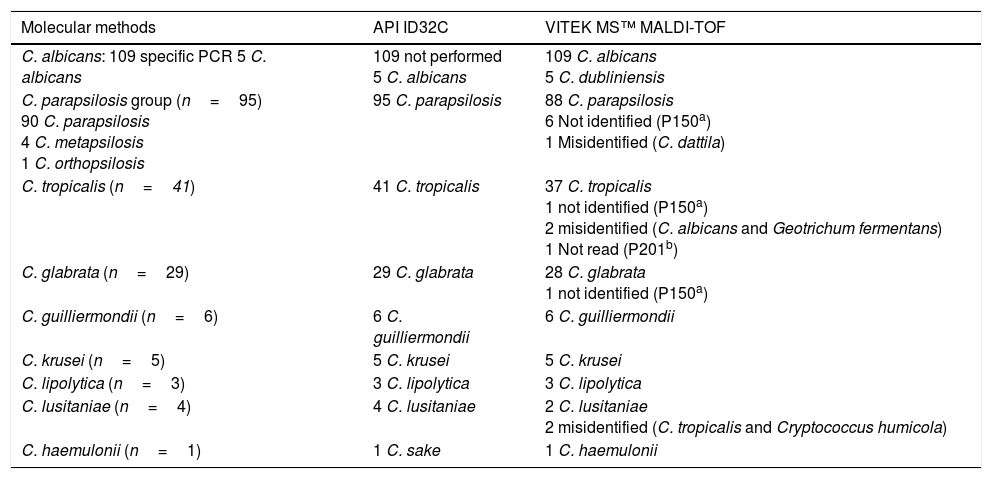
ResultsThe identifications provided for the 298 strains with the indicated conventional method were as follows: 114 C. albicans, 95 Candida parapsilosis, 41 Candida tropicalis, 29 Candida glabrata, 6 Candida guilliermondii, 5 Candida krusei, 4 Candida lusitaniae, 3 Candida lipolytica, 1 Candida haemulonii. The Vitek MS™ system identified correctly to the species level 279 isolates (93.62%); additionally, 10 isolates (3.36%) were misidentified, 8 isolates (2.69%) were not identified and 1 isolate (0.33%) gave a bad spectrum during the acquisition (Table 2).
Identification of Candida species with API ID32C and Vitek MS™.
| Molecular methods | API ID32C | VITEK MS™ MALDI-TOF |
|---|---|---|
| C. albicans: 109 specific PCR 5 C. albicans | 109 not performed 5 C. albicans | 109 C. albicans 5 C. dubliniensis |
| C. parapsilosis group (n=95) 90 C. parapsilosis 4 C. metapsilosis 1 C. orthopsilosis | 95 C. parapsilosis | 88 C. parapsilosis 6 Not identified (P150a) 1 Misidentified (C. dattila) |
| C. tropicalis (n=41) | 41 C. tropicalis | 37 C. tropicalis 1 not identified (P150a) 2 misidentified (C. albicans and Geotrichum fermentans) 1 Not read (P201b) |
| C. glabrata (n=29) | 29 C. glabrata | 28 C. glabrata 1 not identified (P150a) |
| C. guilliermondii (n=6) | 6 C. guilliermondii | 6 C. guilliermondii |
| C. krusei (n=5) | 5 C. krusei | 5 C. krusei |
| C. lipolytica (n=3) | 3 C. lipolytica | 3 C. lipolytica |
| C. lusitaniae (n=4) | 4 C. lusitaniae | 2 C. lusitaniae 2 misidentified (C. tropicalis and Cryptococcus humicola) |
| C. haemulonii (n=1) | 1 C. sake | 1 C. haemulonii |
The Vitek-MS™ system identified 109/114 (95.6%) C. albicans, with the remaining 5 isolates misidentified as Candida dubliniensis. All these 5 isolates were identified as C. albicans by API ID32C, specific PCR and ITS sequencing.
In the “psilosis” group, the Vitek-MS™ gave an identification for 89/95 (93.7%) isolates, with 88 of them (92.6%) correctly identified as C. parapsilosis and one isolate misidentified as Candida dattila. The remaining 6 (6.3%) unidentified isolates in this group were identified by the conventional method as Candida metapsilosis (n=4), Candida orthopsilosis (n=1) or C. parapsilosis (n=1). Both C. metapsilosis and C. orthopsilosis are not included in the database version used in this study.
Vitek-MS™ identified correctly 37/41 (90.2%) C. tropicalis; 2 (4.8%) C. tropicalis were misidentified as C. albicans and Geotrichum fermentans, respectively, another one (2.4%) had no identification and a fourth one (2.4%) gave no read. On CHROMagar Candida plates, 31 (75.6%) strains of C. tropicalis presented colonies with a characteristic blue color at 24h, the remaining 8 (19.5%) presented a cream color at 24h and blue color at 48h, and 2 (4.9%) presented violet colored colonies at both 24 and 48h. That is in agreement with previously published reports.2,18,22
For C. glabrata 28/29 (96.5%) isolates were correctly identified, while 1 (3.5%) isolate was not identified. All C. guilliermondii (n=6), C. krusei (n=5) and C. lipolytica (n=3) were correctly identified by Vitek MS™. Two of four isolates in the C. lusitaniae group were correctly identified by Vitek MS™, while the other 2 (50%) were misidentified as C. tropicalis and Cryptococcus humicola. The only strain of C. haemulonii was correctly identified by Vitek-MS™ but repeatedly misidentified by API-ID32C as C. sake (Table 2).
The Kappa coefficient (κ) between the conventional identification techniques and the MALDI-TOF-MS system was 0.855, which according to the assessment of this index by the Landis and Koch scale, indicates a very good concordance. If we perform this Kappa coefficient (κ) by comparing each of the two methods with the reference method, VitekMS still has a good index (0.91), whereas conventional methods reach almost 1 (0.97).
The sensitivity and specificity of the Vitek-MS™ system reached 82% (IC 95%: 70–90.6%) and 97.2% (IC95%: 94.5–98.8%), respectively, with a positive and negative predictive values of 86.2% (IC95%: 74.60–93.9%) and 96.1% (IC 95%: 93.2–98.1%) respectively. On the other hand, API system sensitivity, specificity, positive predictive value, and negative predictive value were 100% (IC 95%: 92.9–100%), 98% (IC 95%: 95.6–99.2%), 89.3% (IC 95%: 78.1–96%), and 100% (IC 95%: 98.7–100%), respectively. The prevalence of candidemia in our hospital during those years reached only 2%, so there is no influence in those values.
DiscussionThe identification methods in the clinical laboratory should be reliable and fast. The faster diagnosis of MALDI-TOF compared to API method makes the former a good alternative (Kappa coefficient (κ)=0.855), being able to advance the diagnosis 24 or 48h. Our results show an excellent agreement between the conventional identification techniques and the MALDI-TOF-MS system. When comparing the 189 isolates evaluated with these two methods, there was an agreement in the identification of 170 (89.9%) isolates. The API system methodology correctly identified 183 (96.8%) strains while in 6 (3.2%) cases there was no identification to the species level. On the other hand, Vitek MS™ correctly identified 170 (89.9%) strains and did not identify the organism to species level in 19 (10.1%) cases. Our results are within the ranges reported in the literature, perhaps somewhat lower. In our study the number of C. albicans isolates is lower than that of other species of the genus, and no new tests were carried out in spectral failed acquisitions.
In conclusion, the Vitek-MS™ system provides an acceptable and rapid identification at the species level in most Candida species evaluated, showing similar results to other reported studies (Table 1). This is supported by the good data of sensitivity (82%) and specificity (97.2%) obtained. It is a good alternative to conventional methods for the identification of the most frequently isolated yeast species. All these characteristics suggest that the Vitek-MS™ system will have a big impact in patient care and laboratory effectiveness.9 Nevertheless, more studies with a larger number of less frequently isolated Candida species, such as C. lusitaniae, need to be done. Additional studies are also necessary to avoid some cases of misidentification of C. albicans as C. dubliniensis. The errors in the identification between these two phylogenetically closely related yeasts have already been reported in the literature, both with Vitek-MS™ and with BioTyper,8,20 and it would be necessary to propose new mass peaks for the differentiation of their spectra.
Finally, the database of the Vitek-MS™ should include both C. metapsilosis and C. orthopsilosis as they are currently relevant species, not only from the epidemiological point of view, but because of their antifungal resistance profiles.5,7,19 In this regard, other systems such as BioTyper or Saramis (AnagnosTec, Potsdam, Germany and Shimadzu, Duisburg, Germany) have already included these species in their databases.16,21
FundingThis study has been supported by Plan Nacional de I+D+i 2008–2011 and Instituto de Salud Carlos III, Subdirección General de Redes y Centros de Investigación Cooperativa, Ministerio de Economía y Competitividad, Spanish Network for Research in Infectious Diseases (REIPI RD12/0015) – co-financed by European Development Regional Fund “A way to achieve Europe” ERDF.
Conflict of interestsAuthors declare no conflict of interests.