Fungal rhinosinusitis has become an increasingly recognized disease, being Aspergillus species responsible for most of the cases. Its diagnosis is quite difficult because of the non-specific symptoms and low sensitivity of the current diagnostic methods.
AimsAn Aspergillus-specific nested polymerase chain reaction (PCR) assay using biopsy specimens taken from the maxillary sinuses was performed in order to assess its usefulness. Conventional diagnostic methods (histology and culture) were also carried out.
MethodsA case–control study was performed in the Institute of Stomatology, Jagiellonian University in Kraków, between 2011 and 2014. The case group consisted of 21 patients with suspected rhinosinusal mycetoma while the control group included 46 patients with no suspicion of fungal rhinosinusitis. The two-step PCR assay amplified an Aspergillus specific portion of the 18S rRNA gene. Interval estimation of sensitivity, specificity, positive (PPV) and negative (NPV) predictive values were calculated to assess the diagnostic test performance. The agreement between the PCR and the other tests was evaluated using the Kappa coefficient (k).
ResultsNinety percent of the samples obtained from patients diagnosed with mycetoma yielded positive PCR results. The PCR showed almost perfect concordance with histology (k=0.88). Sensitivity, specificity, PPV and NPV estimates were 90%; 95% CI: (55.5–99.7%), 98.3%; 95% CI: (90.9–100%), 90%; 95% CI: (55.5–99.7%) and 98.3%; 95% CI: (90.9–100%), respectively. One clinical sample showed growth of Aspergillus fumigatus and positive PCR despite the negative histological examination.
ConclusionsNested PCR assay is a promising diagnostic tool to evaluate the presence of Aspergillus in the tissue of maxillary sinus from patients with suspicion of sinus aspergillosis.
La rinosinusitis fúngica se ha convertido en una enfermedad cada vez más frecuente y el género Aspergillus es el causante de la mayoría de los casos. Su diagnóstico es relativamente difícil debido a la inespecificidad de los síntomas y a la baja sensibilidad de los métodos de diagnóstico actuales.
ObjetivosEvaluar la eficacia de un ensayo de reacción en cadena de la polimerasa (RCP), con cebadores internos y específica de Aspergillus en muestras de biopsia tomadas de los senos maxilares de algunos pacientes, y compararla con la eficacia de los métodos de diagnóstico convencionales (histología y cultivo).
MétodosSe realizó un estudio de casos y controles en el Instituto de Estomatología de la Universidad Jaguelónica de Cracovia entre 2011 y 2014. El grupo de casos estaba formado por 21 pacientes en que se sospechaba rinosinusitis por micetoma mientras que el grupo control estaba compuesto por 46 pacientes sin sospecha de rinosinusitis fúngica. El ensayo de PCR en dos etapas amplificó una porción específica del gen 18S rRNA de Aspergillus. Se obtuvieron estimaciones de la sensibilidad, la especificidad y de los valores predictivos positivo (VPP) y negativo (VPN) para evaluar el rendimiento de la prueba. La concordancia entre la PCR y las otras pruebas realizadas se evaluó utilizando el coeficiente kappa (k).
ResultadosEl 90% de las muestras obtenidas de pacientes diagnosticados de micetoma mostró resultados positivos en la PCR, con una concordancia casi perfecta de este método con la histología (k=0,88). Las estimaciones de sensibilidad, especificidad, VPP y VPN fueron las siguientes: 90%, IC95% (55,5-99,7%); 98,3%, IC95% (90,9-100%); 90%, IC95% (55,5-99,7%) y 98,3%, IC95%: (90,9-100%), respectivamente. Aspergillus fumigatus se aisló en el cultivo de una muestra clínica, además de obtenerse un resultado positivo por PCR de dicha muestra a pesar de que el examen histológico fue negativo.
ConclusionesEl ensayo de PCR con cebadores internos es una herramienta de diagnóstico prometedora para evaluar la existencia de Aspergillus en tejidos del seno maxilar de pacientes en que se sospeche aspergilosis sinusal.
Rhinosinusitis is an inflammation of the mucous membrane lining, affecting one or more of the paranasal sinuses. The disease may be caused by infectious agents, including viruses, bacteria, and fungi.30 The latter have been increasingly recognized as important pathogens in sinusitis.11,21 Airborne fungi are widely distributed in the human environment. Fungal spores commonly colonize the upper respiratory tract without any adverse effects on human health. However, under certain conditions they may lead to the development of fungal rhinosinusitis (FRS). The disease has been categorized as invasive or noninvasive, based on the histological examination of the tissue that is removed from the sinus. The noninvasive type is characterized by the lack of hyphae within the local tissues of paranasal sinuses and includes fungal colonization, mycetoma (fungus ball) and allergic FRS. The presence of fungal elements within sinus mucosa, submucosa, blood vessels, or bone indicates invasive form of FRS.26 Pathogens responsible for FRS may differ according to the type of sinusitis and the geographical prevalence of certain fungal species; however it has been reported that Aspergillus is one of the leading causes of FRS.12,15,18 Early recognition and accurate classification of sinus aspergillosis are crucial to give an effective therapy and achieve a better prognosis.14 Diagnostic process involves many different and confusing aspects. Histology is considered to be the gold standard, but the identification of the species is not possible. While fungal culture allows the identification of the aetiological agents, it has low sensitivity.4 For these reasons it is necessary to improve the diagnostic methods for sinus aspergillosis.
There are a several reports about molecular identification of Aspergillus DNA in the tissue samples by using different approaches of DNA-amplification, e.g. real-time quantitative and traditional PCR assay targeting Aspergillus mitochondrial DNA followed by products sequencing, PCR-restriction fragment length polymorphism analysis, or multiplex PCR combined with consecutive DNA microarray hybridization.2,3,6,16,23,24,27 Nested PCR assay, proposed by some authors, has shown to be highly efficient in the identification of Aspergillus species, but its ability to diagnose sinus aspergillosis has not been studied.22,23,25 Therefore, the aim of this study was to optimize a molecular assay for detecting Aspergillus in the biopsy specimens of maxillary sinus mucosa and to compare the efficacy of this method with that of histological examination and culture in the diagnosis of rhinosinusitis caused by Aspergillus species.
Materials and methodsPatients and clinical samplesThe study material consisted of 69 biopsy samples of maxillary sinus mucosa taken intraoperatively from 67 patients treated for chronic sinusitis in the Department of Cranio-Maxillofacial, Oncological and Reconstructive Surgery of the Institute of Stomatology, Jagiellonian University in Kraków in the period from 2011 to 2014.
All the patients underwent the Caldwell-Luc procedure. One patient underwent a subsequent revision endoscopic sinus surgery. Tissue sampling was performed as clinically indicated, not solely for the purposes of the study. Twenty-one patients (16 women and 5 men, within the age range of 21–67 years, average age of 38) were diagnosed with suspected mycetoma according to the following criteria: prolonged and ineffective antibiotic therapy confirmed by interview, presence of a “metallic tone” intrasinusally, presence of foreign body-like structures visible on computed tomography scans, and tissue congestion or chalk-like concretions found within the sinus during surgery. In this group, one patient (PXIII) underwent Caldwell-Luc surgery twice in two months, and then, after a year, the patient had a revision endoscopic sinus surgery. During these medical procedures three clinical samples were obtained from this patient. The total number of samples in the group of patients with suspicion of mycetoma was 23. The remaining 46 patients (22 women and 24 men, within the age range of 19–79 years, average age of 40) with no clinical suspicion of FRS were included in the study as the control group. From these patients a total of 46 specimens were collected. The study was approved by the Ethics Committee of the Jagiellonian University Medical College (KBET/129/B/2011). Written consent was obtained from each patient.
Analysis of clinical specimensAnalysis of clinical specimens included histological and microbiological examinations performed in the Department of Laboratory Diagnostics of Ludwik Rydygier Memorial Hospital. The molecular tests were performed in the Department of Pharmaceutical Microbiology of the Jagiellonian University Medical College. Histopathological staining of the specimens was carried out using haematoxylin-eosin (HE), periodic acid Schiff (PAS) or Gomori-Grocott stain. Microbiological analysis was based on culture on Sabouraud dextrose agar (bioMérieux, Marcy l’Etoile, France) supplemented with chloramphenicol and incubated at 30 and 37°C for 10 days.14,18 The identification of the moulds was performed under microscopic examination of lactophenol cotton blue preparation. The identification of the yeast isolates was achieved by means of the automatic Vitek 2 Compact system (bioMérieux). Mucosal biopsy specimens were stored in sterile tubes at −80°C for molecular purposes.
DNA preparationThe biopsy specimens weighing <20mg were used for DNA extraction. The following procedures were conducted: homogenization of the tissue in liquid nitrogen, initial enzymatic digestion with litycase (Sigma–Aldrich, Darmstadt, Germany), and DNA purification using a commercial spin-column-based GeneMATRIX Plant&Fungi DNA Purification Kit (EURx, Gdańsk, Poland).
PCR reactionsIn the nested PCR reactions, two pairs of primers amplifying the gene fragment encoding the 18S rRNA subunit, described previously by Skladny et al., were used to detect Aspergillus species.25 The outer set of primers, AFU7S and AFU7AS, produces a fragment of 405-bp, and the inner set of primers, AFU5S and AFU5AS, generates a 236-bp product.
The first round of the nested PCR reaction was performed with 20–40ng of DNA, whereas in the second round of amplification, 2μl of PCR product from the first reaction were used. Each PCR contained 1× GoTaq Flexi Buffer (pH 8.5) (Promega, Madison, Wisconsin, USA), 2mM MgCl2, 0.4μM of primers (outer for the first step and inner for the second step) (Blirt, Gdańsk, Poland), 0.2mM of each nucleotide (Thermo Scientific, Waltham, Massachusetts, USA), and 0.625U of GoTaq DNA Polymerase (Promega). The PCR reactions were performed in a thermocycler T personal (Biometra, Göttingen, Germany). The first round of PCR consisted of initial denaturation at 95°C for 2min, 40 cycles of denaturation at 94°C for 40s, annealing at 65°C for 1min, elongation at 72°C for 1min, and a final extension at 72°C for 5min. The second round of amplification consisted of initial denaturation at 95°C for 2min and 35 cycles carried out under the same thermal conditions as the first round of nested PCR, followed by a final extension step of 5min at 72°C. PCR amplification was performed twice for each sample. The control samples included DNA of Aspergillus fumigatus ATCC 14110 reference strain as a positive control and two negative controls: the first control without the addition of DNA and the second one containing DNA isolated from the human colonic adenocarcinoma cell line Caco-2.
The DNA fragments were separated by 2% agarose gel electrophoresis and visualized by ethidium bromide staining. The PCR product size was determined by comparison with a molecular weight standard (O’Gene Ruler 100bp DNA Ladder Plus; Thermo Scientific). A 138-bp gene fragment of the human glucose-6-phosphate dehydrogenase was used as an internal control.25
Species specificity and detection limit of PCRThe species specificity of the PCR assay was tested using reference and clinical strains of bacteria and fungi, most often isolated from the biopsy samples of maxillary sinus mucosa from patients treated in the Department of Cranio-Maxillofacial, Oncological and Reconstructive Surgery of the Institute of Stomatology, Jagiellonian University, Kraków. These species included Staphylococcus aureus ATCC 25923, Staphylococcus epidermidis – clinical strain, Escherichia coli ATCC 25922, Klebsiella pneumoniae ATCC 700603, Enterobacter cloacae Microbank 10.011, Proteus mirabilis – clinical strain, Acinetobacter baumannii ATCC 19606, Candida albicans ATCC 90028, Candida glabrata ATCC 2001, Candida dubliniensis – clinical strain, A. fumigatus ATCC 14110, Aspergillus flavus – clinical strain and Aspergillus niger – clinical strain.
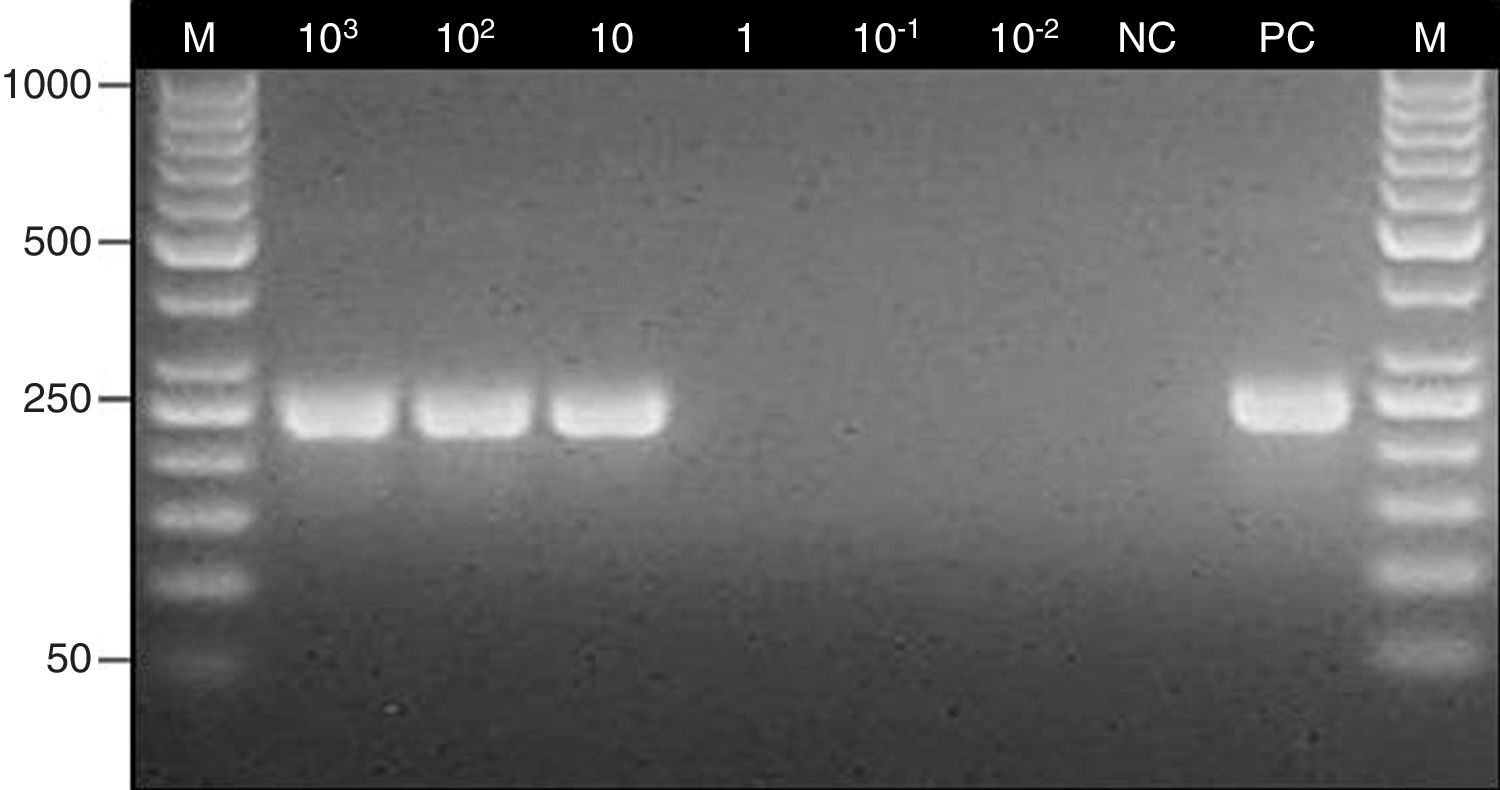
The detection limit of the method was determined by performing PCR assays with six 10-mg weighted fragments of the maxillary sinus mucosal biopsy specimen taken from a patients in whom mycetoma was not suspected; the specimens were spiked with different quantities of A. fumigatus ATCC 14110 conidia (from 103 to 10−2). A fragment of a biopsy specimen of the maxillary sinus mucosa with no conidia was used as negative control.
Statistical analysisData was analyzed using PQStat software (1.6.4 version). Only those cases that were positive by histological findings were considered true positive. Statistical analysis of agreement between PCR and histology was performed using the Cohen's Kappa test, with the Kappa values (k) ranging from −1 to 1, where “1” was considered as “perfect” agreement between tests, and ranges from 0.81 to 0.99, 0.61 to 0.8, 0.41 to 0.6, 0.21 to 0.4, and 0.0 to 0.2 were considered the “almost perfect”, “substantial”, “moderate”, “fair” and “slight” agreement, respectively. Measures of diagnostic efficacy (sensitivity, specificity, positive (PPV) and negative (NPV) predictive values, and 95% confidence intervals (CI)) were calculated using histological examination as the gold standard. P-Values less than 0.05 were considered statistically significant.
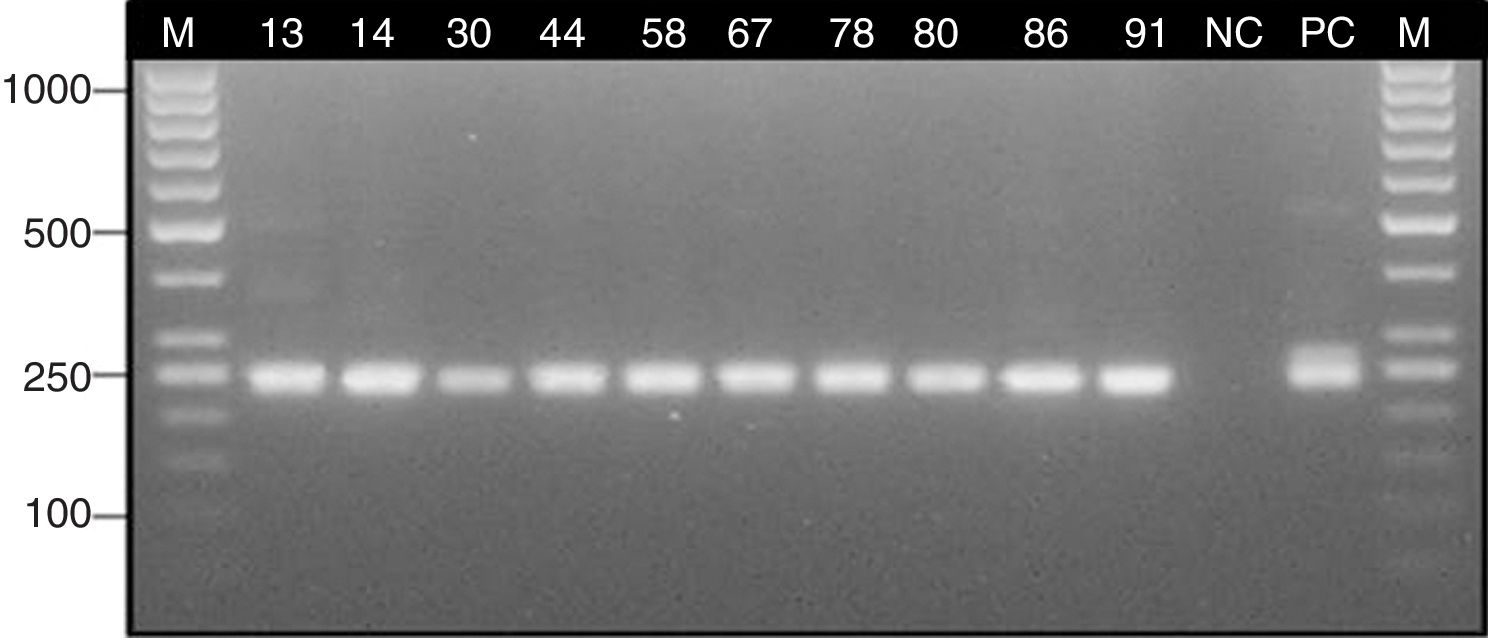
ResultsStudy group – patients with suspicion of sinus mycetomaSeveral Aspergillus isolates (six A. fumigatus, and two Aspergillus candidus) were obtained in culture from six clinical samples (26.1%) of six different patients with suspicion of mycetoma (28.6%). Two isolates of Candida albicans from two other patients were also obtained (9.5%) (Table 1). Histological staining indicated the presence of septate hyphae with dichotomous branching at 45° in 10 clinical specimens (43.5%) obtained from eight patients with suspicion of mycetoma (38.1%) (Table 1). Nested PCR assay confirmed the presence of Aspergillus DNA in eight patients with suspicion of mycetoma (38.1%) (Table 1). The expected ∼240 bp fragment of 18S rRNA gene was detected in 10 clinical samples (43.5%) (Fig. 3).
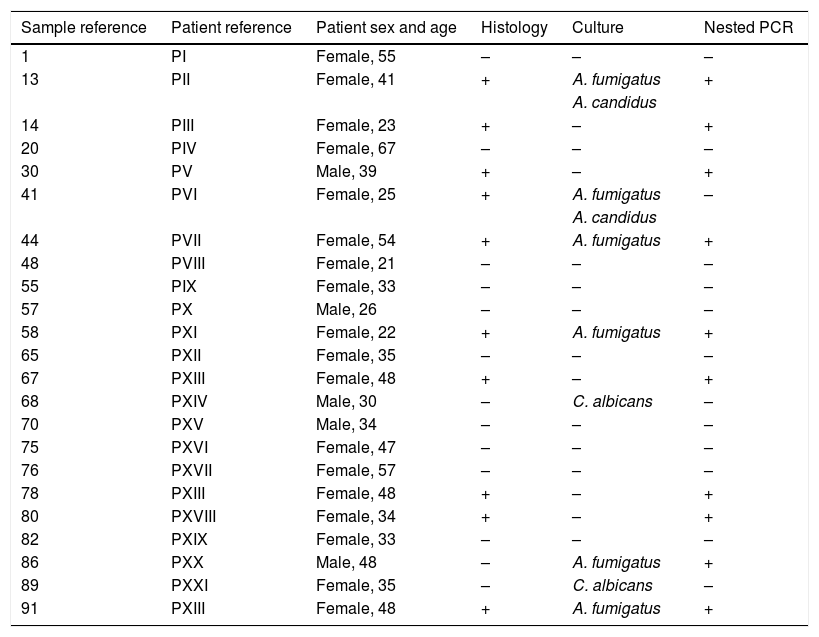
Comparison of the nested PCR, culture and histological results in the patients with suspicion of sinus mycetoma.
| Sample reference | Patient reference | Patient sex and age | Histology | Culture | Nested PCR |
|---|---|---|---|---|---|
| 1 | PI | Female, 55 | – | – | – |
| 13 | PII | Female, 41 | + | A. fumigatus | + |
| A. candidus | |||||
| 14 | PIII | Female, 23 | + | – | + |
| 20 | PIV | Female, 67 | – | – | – |
| 30 | PV | Male, 39 | + | – | + |
| 41 | PVI | Female, 25 | + | A. fumigatus | – |
| A. candidus | |||||
| 44 | PVII | Female, 54 | + | A. fumigatus | + |
| 48 | PVIII | Female, 21 | – | – | – |
| 55 | PIX | Female, 33 | – | – | – |
| 57 | PX | Male, 26 | – | – | – |
| 58 | PXI | Female, 22 | + | A. fumigatus | + |
| 65 | PXII | Female, 35 | – | – | – |
| 67 | PXIII | Female, 48 | + | – | + |
| 68 | PXIV | Male, 30 | – | C. albicans | – |
| 70 | PXV | Male, 34 | – | – | – |
| 75 | PXVI | Female, 47 | – | – | – |
| 76 | PXVII | Female, 57 | – | – | – |
| 78 | PXIII | Female, 48 | + | – | + |
| 80 | PXVIII | Female, 34 | + | – | + |
| 82 | PXIX | Female, 33 | – | – | – |
| 86 | PXX | Male, 48 | – | A. fumigatus | + |
| 89 | PXXI | Female, 35 | – | C. albicans | – |
| 91 | PXIII | Female, 48 | + | A. fumigatus | + |
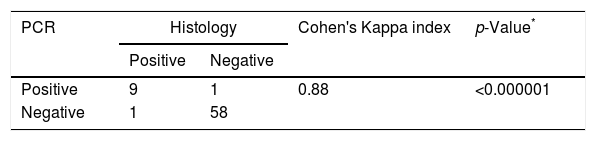
The results of PCR assay were consistent with the histological diagnosis in 9 out of 10 samples. The presence of Aspergillus could not be confirmed by the molecular method only in the tissue fragment no. 41, but it is worth noting that this clinical sample was positive by culture. On the other hand, in one tissue biopsy specimen (no. 86), Aspergillus was not identified by histological staining while the other methods (PCR and culture) confirmed the presence of this mould (Table 1). The agreement (Cohen's Kappa coefficient) between the results of PCR and the histological findings in tissue biopsies was quite high (k=0.88) (Table 2).
Agreement between PCR and histology findings in the diagnosis of sinus mycetoma (n=69).
| PCR | Histology | Cohen's Kappa index | p-Value* | |
|---|---|---|---|---|
| Positive | Negative | |||
| Positive | 9 | 1 | 0.88 | <0.000001 |
| Negative | 1 | 58 | ||
One A. fumigatus strain was isolated in the culture of the sample collected from one patient in the control group (2.2%); however the lack of clinical and histological evidences indicated that its presence in culture was due to contamination. Additionally six Candida strains were isolated from five patients (10.9%): four C. albicans, one C. dubliniensis and one Candida kefyr (13%) (Table 3). None of the patients without suspicion of mycetoma had a positive histological or nested PCR results.
Comparison of the nested PCR, culture and histological results in the patients without suspicion of sinus mycetoma.
| Sample reference | Patient reference | Patient sex and age | Histology | Culture | Nested PCR |
|---|---|---|---|---|---|
| 6 | P23 | Female, 32 | – | C. dubliniensis | – |
| 7 | P24 | Male, 29 | – | – | – |
| 8 | P25 | Male, 42 | – | – | – |
| 10 | P26 | Male, 26 | – | – | – |
| 11 | P27 | Male, 47 | – | – | – |
| 15 | P28 | Male, 62 | – | – | – |
| 16 | P29 | Male, 36 | – | – | – |
| 17 | P30 | Male, 22 | – | – | – |
| 18 | P31 | Male, 35 | – | – | – |
| 19 | P32 | Male, 38 | – | – | – |
| 21 | P33 | Male, 19 | – | – | – |
| 24 | P34 | Female, 42 | – | – | – |
| 25 | P35 | Female, 57 | – | – | – |
| 26 | P36 | Female, 57 | – | – | – |
| 29 | P37 | Female, 40 | – | – | – |
| 31 | P38 | Female, 35 | – | – | – |
| 32 | P39 | Female, 36 | – | – | – |
| 33 | P40 | Female, 27 | – | – | – |
| 34 | P41 | Female, 44 | – | – | – |
| 36 | P42 | Male, 79 | – | C. albicans / C. kefyr | – |
| 37 | P43 | Male, 33 | – | – | – |
| 39 | P44 | Female, 39 | – | – | – |
| 42 | P45 | Female, 41 | – | – | – |
| 43 | P46 | Male, 52 | – | – | – |
| 45 | P47 | Male, 37 | – | – | – |
| 46 | P48 | Female, 41 | – | C. albicans | – |
| 47 | P49 | Male, 31 | – | – | – |
| 49 | P50 | Female, 31 | – | – | – |
| 50 | P51 | Male, 41 | – | – | – |
| 51 | P52 | Female, 57 | – | – | – |
| 59 | P53 | Male, 39 | – | C. albicans | – |
| 60 | P54 | Male, 48 | – | – | – |
| 61 | P55 | Male, 36 | – | – | – |
| 62 | P56 | Male, 42 | – | – | – |
| 64 | P57 | Male, 42 | – | – | – |
| 71 | P58 | Male, 58 | – | – | – |
| 72 | P59 | Female, 31 | – | C. albicans | – |
| 73 | P60 | Female, 31 | – | – | – |
| 77 | P61 | Male, 32 | – | – | – |
| 79 | P62 | Female, 66 | – | – | – |
| 84 | P63 | Female, 21 | – | – | – |
| 85 | P64 | Female, 44 | – | A. fumigatusa | – |
| 87 | P65 | Female, 19 | – | – | – |
| 88 | P66 | Female, 39 | – | – | – |
| 90 | P67 | Female, 31 | – | – | – |
| 92 | P68 | Male, 35 | – | – | – |
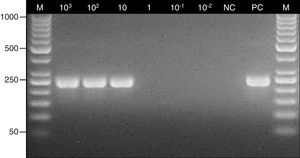
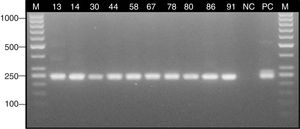
The PCR assay was almost specific for Aspergillus species since the specific products were obtained only with A. fumigatus, A. flavus and A. niger DNA, whereas PCRs with the other fungal and bacterial strains were negative (Fig. 1). The limit of detection of the method was 10 conidia/10mg of tissue (1CFU/mg) (Fig. 2).
Determination of the specificity of the nested PCR assay against reference and clinical strains of bacteria and fungi. Lane: M, 100-bp molecular size marker (Thermo Scientific); AF, Aspergillus fumigatus ATCC 14110; AFL, Aspergillus flavus – clinical isolate; AN, Aspergillus niger – clinical isolate; CA, Candida albicans ATCC 90028; CG, Candida glabrata ATCC 2001; CD, Candida dubliniensis – clinical isolate; SA, Staphylococcus aureus ATCC 25923; SE, Staphylococcus epidermidis – clinical isolate; EC, Escherichia coli ATCC 25922; KP, Klebsiella pneumoniae ATCC 700603; ECL, Enterobacter cloacae Microbank 10.011; PM, Proteus mirabilis – clinical isolate; AB, Acinetobacter baumannii ATCC 19606; NC1, negative control (DNA isolated from the human colonic adenocarcinoma cell line Caco-2); NC2, negative control (nuclease-free water).
Determination of the detection limit for the nested PCR assay. Lane: M, 100-bp molecular size marker (Thermo Scientific); 103–10−2, fragments of maxillary sinus mucosal biopsy spiked with several quantities of Aspergillus fumigatus ATCC 14110 conidia (from 103 to 10−2); NC, negative control; PC, positive control (Aspergillus fumigatus ATCC 14110).
Sensitivity, specificity, PPV, NPV and 95% CI of the nested PCR and culture are presented in Table 4 in comparison with the histological test (gold standard for the diagnosis of FRS). The prevalence of sinus aspergillosis estimated in this study on the basis of the histological examination of the tissues was 14% (10/69).
Diagnostic parameters of the nested PCR and culture compared with the reference histological test (with the study prevalence of 14%).
Over the past two decades, FRS has been increasingly diagnosed in patients with chronic rhinosinusitis. Aspergillus species are the predominant aetiological agents of FRS, with the exception of the acute invasive form most frequently caused by zygomycetes.10,15,18 The primary species involved in FRS are A. fumigatus and A. flavus, and rarely Aspergillus nidulans, Aspergillus terreus, Aspergillus oryzae or Aspergillus avenaceus.5,17 In accordance with the already-mentioned reports, A. fumigatus has been the most commonly isolated species in our study.
Achieve the diagnosis of sinus aspergillosis is often difficult. The most important step, which often brings the correct diagnosis, is the histological observation of 45° angle dichotomous branching and septate hyphae in the sinus tissues. However, many other rarely isolated fungi, such as Scedosporium or Fusarium, may present similar structures.8 Mycological examination based on culture is also a key element in the diagnostic process as the moulds can be identified to the species level.20 However this method has also drawbacks due to its low sensitivity, which has reached 50% (95% CI 18.7–81.3%) in our study. Our findings are in accordance with the observations of other authors, in which the culture sensitivity values were 34.5–61.7%, depending on the classification of FRS.7,10,14,19
Due to the disadvantages of the above-mentioned methods, molecular approaches can be a useful tool in the diagnosis of FRS. A variety of these techniques to detect different fungi in human samples have already been reported. Willinger et al. used a PCR assay targeting fungal 28S ribosomal DNA in fresh and paraffin-embedded tissues sections of the maxillary sinus of patients with histologically proven fungus balls. The authors found that PCR is highly sensitive in comparison with histology, reaching 100% in fresh tissues and 87.7% in paraffin-embedded samples. They also revealed a higher sensitivity of PCR when compared to culture.29 On the other hand, Kostamo et al. found that using the A. fumigatus-specific PCR method did not increase the accuracy of FRS diagnosis.13
Several studies have shown the high sensitivity and specificity of nested PCR with the primers AFU5S, AFU5AS, AFU7S and AFU7AS in the diagnosis of invasive aspergillosis (IA). Hummel et al., who detected Aspergillus DNA in blood, CSF and BAL of children and adolescent patients with proven/probable IA, found that the sensitivity and specificity was 80% and 81%, respectively.9 Other studies reported a sensitivity of 89–100% and a specificity of 93–100% in fresh tissue taken by biopsy or in CSF from individuals with proven/probable IA.22,23 Also Teifoori et al. confirmed the usefulness of this method to detect Aspergillus DNA in the blood of bone marrow transplantation patients.28 These findings were in accordance with the results of our study in which the sensitivity and specificity of the nested PCR were 90% (95%, CI 55.5–99.7%) and 98.3% (95%, CI 90.9–100%), respectively. However, Badiee et al., who examined the functional endoscopic surgery specimens obtained from patients with rhinosinusitis, found that the specificity of this method was only 28.2%. It should be emphasized, however, that these authors used other criteria to diagnose a fungal infection of the sinuses; they regarded a positive fungal culture as sufficient proof of infection.1
It should be noted that all statistical parameters of the nested PCR assay, included sensitivity, specificity, positive and negative predictive values, were calculated with a value of 14% in sinus aspergillosis prevalence (the percentage established in this study). While the sensitivity and specificity are intrinsic attributes of the diagnostic test, positive and negative predictive values are highly dependent on the prevalence of the disease in the population tested. According to the literature, sinus mycetoma is encountered in about 13–28.5% of the chronic rhinosinusitis cases and the fungi most often obtained in culture are Aspergillus species.8 However, the incidence and aetiological agents of certain particular types of FRS varies among different geographical areas.14,17
The efficacy of the molecular methods to detect directly fungal DNA in human samples is highly dependent on the type of the clinical specimens and the DNA extraction protocol. It has been proved that fresh or frozen tissues offer a better chance than paraffin-embedded specimens burdened with the risk of the DNA degradation.3 The method optimized in this study demonstrated a detection limit of about 1CFU per mg of tissue. These results have confirmed that the nested PCR can detect even small amounts of fungus present in the sinus tissue sections. The high sensitivity and specificity of the method were previously described by Skladny et al., who introduced this assay to detect Aspergillus fungi in blood. The authors obtained the detection threshold of the PCR assay between 1 and 5 CFU per ml of blood (100% specificity), determined with 35 bacterial and fungal strains, including Penicillium, Paecilomyces variotii, Rhizopus oryzae and Fusarium proliferatum.25
In conclusion, the proper species identification is of great relevance in the progress of patients with fungal rhinosinusitis. Early recognition and classification of the disease enables to start an appropriate therapy, which is a key step in the patient management. In the last years several molecular techniques have been developed to improve the diagnosis of aspergillosis. While DNA sequencing of the fungal rDNA ITS region amplified by qPCR is considered the reference method of the species confirmation, nested PCR assay, described in this study, has proved to be a valuable tool in the correct identification of Aspergillus. Although nested PCR has the limitation of cross contaminations, the advantages of this method, including simplicity, time and cost-effectiveness, and accuracy, makes it a useful method in the diagnosis of fungal infections, especially when the advanced technology is not available and when the sequencing of the products is not possible. The high sensitivity and specificity of the nested PCR assay reported in this study show its suitability in the diagnosis of sinus aspergillosis.
Authors’ declarationScientific work was supported by the Jagiellonian University Medical College with founds from maintenance of the research potential of the JU MC Department of Pharmaceutical Microbiology.
Conflict of interestThe authors declare that there are no conflicts of interest.
We would like to thank Piotr Pierzchalski for kindly providing the Caco-2 cell line used in this study.