HER2-positive tumors is one of the aggressive subtypes of breast cancer that indicate bad prognosis. Trastuzumab is one of the targeted therapy which inhibit HER2 receptors. Mutations/expression deregulations of the downstream of HER2 receptors could cause resistance to trastuzumab. PTEN is a tumor suppressor gene which directly regulates PI3K pathway which renders it one of the predictive markers of trastuzumab.
MethodsIn the present study, PTEN mutations were screened in 51 patients with HER2-positive breast cancer. Also, 16 patients were further analyzed for protein expression.
ResultsThe mutations were detected in 3 out of 51 patients (5.9%). In addition, 56.3% of the 16 patients showed downregulation/loss of PTEN protein expression. The loss of PTEN was found in 75% of estrogen-receptor negative patients (p =0.130).
ConclusionsThe downregulation/loss of PTEN protein has the tendency to be associated with ER-negative reflecting its value as a treatment prediction marker.
Los tumores HER2 positivos son uno de los subtipos agresivos de cáncer de mama que indican un mal pronóstico. Trastuzumab es una de las terapias dirigidas que inhiben los receptores HER2. Las mutaciones/desregulaciones de la expresión aguas abajo de los receptores HER2 podrían causar resistencia a trastuzumab. PTEN es un gen supresor de tumores que regula directamente la vía PI3K, lo que lo convierte en uno de los marcadores predictivos de trastuzumab.
MetodosEn el presente estudio, las mutaciones de PTEN se examinaron en cincuenta y un pacientes con cáncer de mama positivo para HER2. Además, dieciséis pacientes fueron analizados más a fondo para determinar la expresión de proteínas.
ResultadosLas mutaciones se detectaron en tres de 51 pacientes (5.9%). Además, el 56,3 % de los dieciséis pacientes mostró regulación baja/pérdida de la expresión de la proteína PTEN. También se encontró pérdida de PTEN en el 75% de los pacientes con receptores de estrógeno negativos.
ConclusionesLa regulación baja/pérdida de la proteína PTEN tiende a asociarse con ER-negativo, lo que refleja su valor como marcador de predicción de tratamiento.
Overexpression of Her-2 (Human epidermal growth factor receptor 2) is found in approximately 25% of human breast cancers leading to an aggressive phenotype and poor patient survival. HER2 is a tyrosine kinase receptor; in invasive breast carcinomas, it is overexpressed or amplified in 20–30% of patients.1
Overexpression of HER2 protein could cause receptor homo/heterodimerization which in turn induces the phosphorylation of the intracellular domain and subsequently downstream signaling including PI3K/AKT. Activated PI3K catalyzes the phosphorylation of inositol lipids to produce phosphatidylinositol-3, -4, -5 trisphosphate (PIP3), which is dephosphorylated to PIP2 by phosphatase and tensin homolog (PTEN). In the downstream signaling, PIP3 activates the serine/threonine kinase AKT which in turn regulates the mammalian target of rapamycin (mTOR).2
PTEN is a tumor suppressor gene which is commonly inactivated by many mechanisms in several sporadic cancers.3,4 It is a lipid phosphatase which inhibits PI3K pathway. In sporadic breast cancer, PTEN is mutated in approximately 5% of patients. On the expression level, approximately 25% of breast cancer cases.5,6
Since mutations in downstream genes of HER2 could interfere with trastuzumab treatment, studying mutations/protein expression of these genes could have clinical impact to predict the response to the expensive targeted therapy. So, our present study aimed to screen PTEN mutations as well as loss of protein in HER2-positive breast cancer patients.
Materials and methodsPatients and samplesThe samples were recruited from the National Cancer Institute, Cairo University in collaboration with the Early Cancer Detection Unit, Faculty of Medicine, Ain Shams University Maternity Hospital. The study was approved by the ethics committee of the National Cancer Institute (IRB2010012036B.1).
Formalin fixed paraffin embedded (FFPE) sections were collected from 51 female patients, who were diagnosed with HER2 positive breast cancer between 2007 and 2013. Also, clinical and pathological information, including age, tumor type, tumor grade, marital and menopausal status, lymph nodes, HER2 score and ER and PR status, were collected. Also, PIK3CA mutations were reported earlier for this cohort.7 HER2 score is determined by immunohistochemistry and in some cases FISH analysis is also done.
Genomic DNA was purified from FFPE tissues using QIAamp DNA extraction kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol with slight modifications. DNA concentration and purity was quantitated using the Nanodrop spectrophotometer.
Detection of mutations for PTEN geneIn this study, PTEN mutations were detected in Exons 5, 6, and 7 where most of hotspots are known. PCR amplification was used using the following primers from previous publications8–10: exon5 F (GCAACATTTCTAAAGTTACCTA) and exon5 R (CTGTTTTCCAATAAATTCTCA), exon6 F (CATAGCAATTTAGTGAAATAACT) and exon6 R (GATATGGTTAAGAAAACTGTTC), and exon7 F (CAGTTAAAGGCATTTCCTGTG) and exon7 R (GGATATTTCTCCCAATGAAAG). The PCR products were sequenced by Sanger sequencing and all sequencing reactions were performed using both primers from different direction.
The resulted sequences were aligned to human genomic DNA sequence that present in the GenBank using BLAST software. Mutations and SNPs identification numbers were retrieved using ENSEMBL and COSMIC (Catalogue of Somatic Mutations in Cancer) databases.
Evaluation of PTEN protein expression by Immunohistochemistry (IHC)For 16 patients, IHC was done to assess the protein expression in these samples. For each formalin-fixed and paraffin-embedded tumors, 4-μm-thick section was processed for immunostaining (Santa Cruz, CA, USA). Blocking was done with 3% hydrogen peroxide for 10 min and was washed with TBS to evade Endogenous peroxidase activity of the tissue. Then, serum blocking was performed for 10 min. Subsequently, sections were incubated overnight at room temperature with Human/Mouse/Rat PTEN antibody monoclonal mouse IgG1 Catalog # sc-7974 from Santa Cruz Biotechnology. After two washing steps in TBS, slides were incubated with 100 μl of horseradish peroxidase (HRP) labeled polymer Rabbit/Mouse for 10 min at room temperature and washed in TBS. Then, slides were treated with 3,3-Diaminobenzidine (DAB) + substrate/chromogen solution for 5–10 min and washed in ddH2O. For counterstaining, sections were stained with Mayer's hematoxylin, rinsed in tap water, dehydrated, cleared and cover-slipped.
Internal normal tissues were used as positive controls. Immunohistochemical reactivity was graded, according to the percentage of positive tumor cells: -, 0; +, <20%; ++, 20–50%; and +++, >50%. Grade (-) or (+) was considered as negative expression and grades (++) and (+++) were considered as positive expression.
Statistical analysisStatistical analysis was executed using categorical testing (Chi-square and Fisher's exact tests), in order to determine the relation between qualitative variables. p-value ≤0.05 was considered significant. All tests were two tailed.
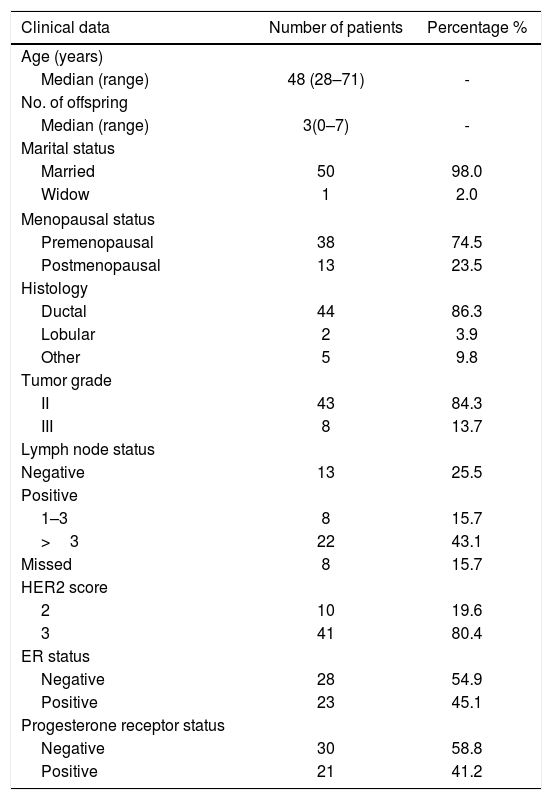
ResultsClinical characteristics of the study cohortFormalin-fixed paraffin embedded tissues of 51 HER2-positive patients were recruited in this study from the National Cancer Institute, Cairo University in collaboration with the Early Cancer Detection Unit, Faculty of Medicine, Ain Shams University Maternity Hospital. According to IHC and/or FISH analysis done as a routine for breast cancer patients, 10 patients were with HER2 score 2 (19.6%) and 41 patients (80.4%) with score 3. The cohort age at time of sample collection ranged from 28 to 71 years old with median age of 48. Estrogen receptor status was positive in 23 patients (45.1%) and negative in 28 cases (54.9%). According to histology, the studied cohort is classified to ductal carcinoma (86.3%), lobular (3.9%), and other classification (9.8%). All available clinical characteristics of patients are shown in Table 1.
Clinical characteristics of the study cohort.
| Clinical data | Number of patients | Percentage % |
|---|---|---|
| Age (years) | ||
| Median (range) | 48 (28–71) | - |
| No. of offspring | ||
| Median (range) | 3(0–7) | - |
| Marital status | ||
| Married | 50 | 98.0 |
| Widow | 1 | 2.0 |
| Menopausal status | ||
| Premenopausal | 38 | 74.5 |
| Postmenopausal | 13 | 23.5 |
| Histology | ||
| Ductal | 44 | 86.3 |
| Lobular | 2 | 3.9 |
| Other | 5 | 9.8 |
| Tumor grade | ||
| II | 43 | 84.3 |
| III | 8 | 13.7 |
| Lymph node status | ||
| Negative | 13 | 25.5 |
| Positive | ||
| 1–3 | 8 | 15.7 |
| >3 | 22 | 43.1 |
| Missed | 8 | 15.7 |
| HER2 score | ||
| 2 | 10 | 19.6 |
| 3 | 41 | 80.4 |
| ER status | ||
| Negative | 28 | 54.9 |
| Positive | 23 | 45.1 |
| Progesterone receptor status | ||
| Negative | 30 | 58.8 |
| Positive | 21 | 41.2 |
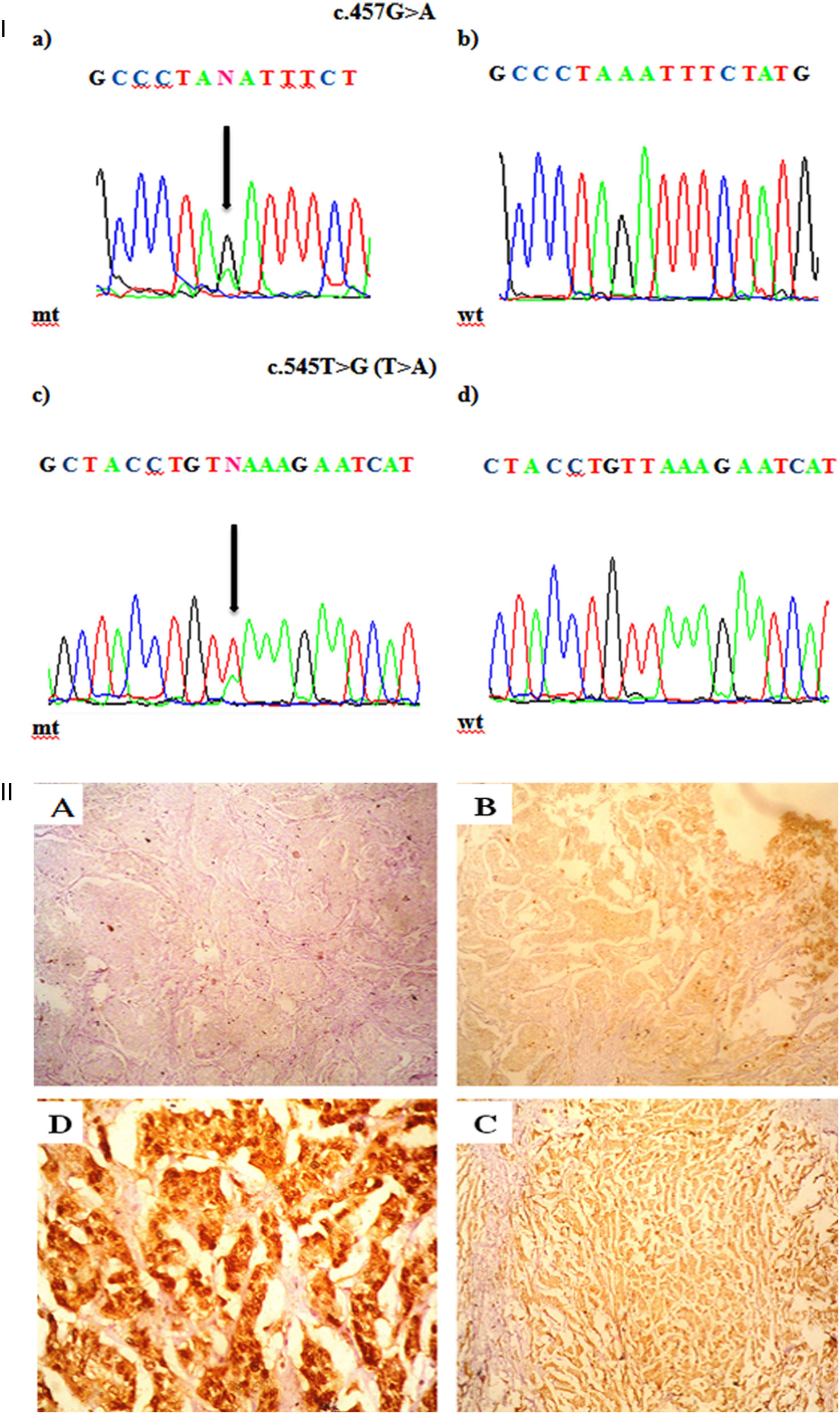
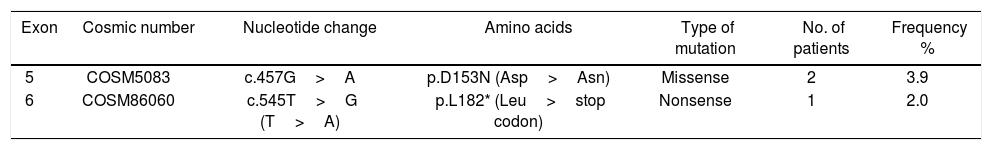
Mutations were investigated in exons -5, -6, and -7. Three out of 51 patients (5.8%) harbored mutations in PTEN, in which two patients harbored one missense mutation and one has nonsense mutation. All detected mutations were heterozygous. The types of mutations found are described in Table 2 & Fig. 1a; where codon 457 G>A missense mutation leading to an amino acid change from Aspartic Acid to Asparagine (D153N) is found in two patients. The second mutation was nonsense mutation in exon 6 that create stop codon (L182*). No mutation was detected in exon 7.
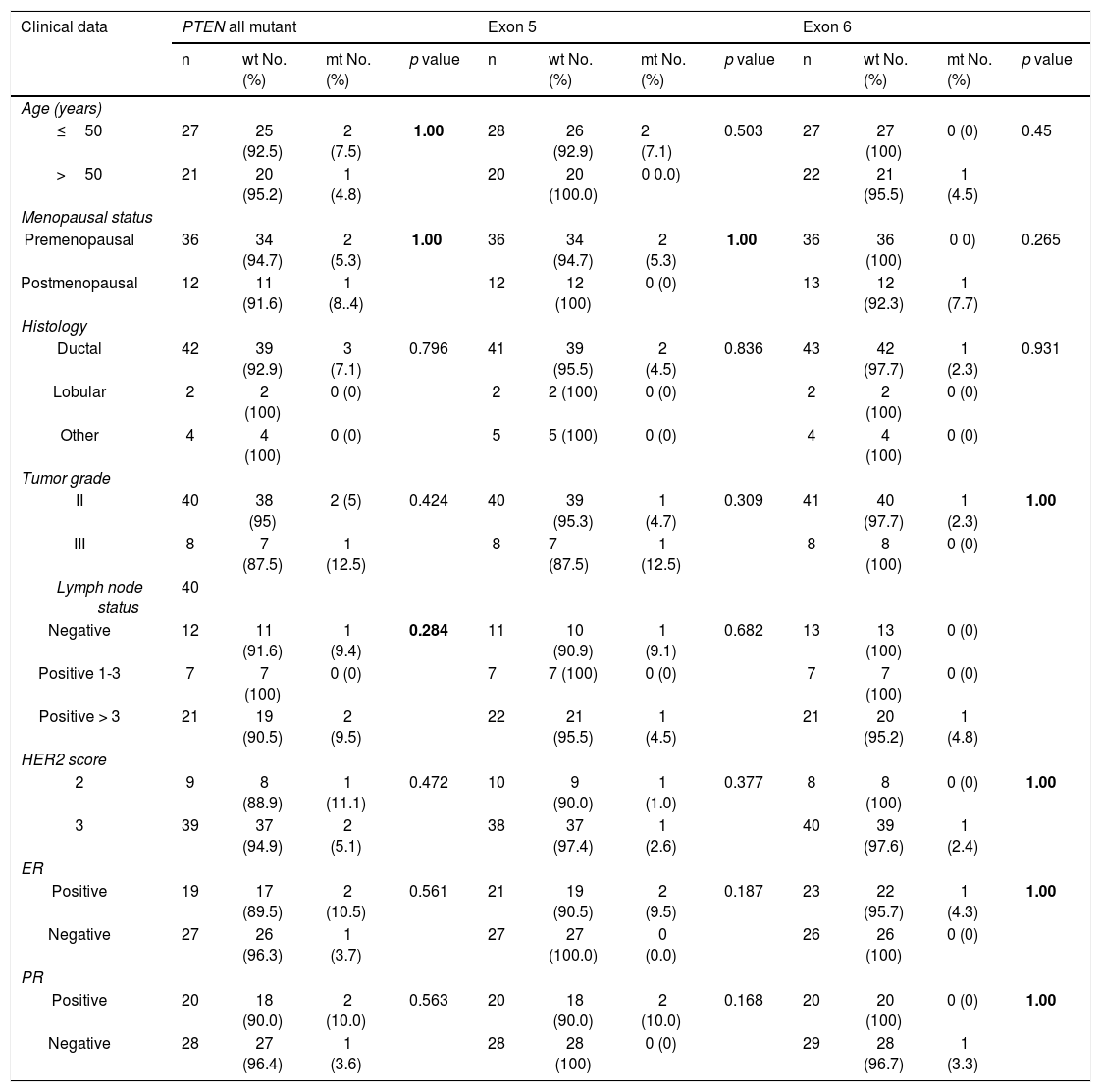
I. a. Partial chromatograms of PTEN mutations: a and c, mutations (457G>A and 545T>G (T>A)) of exon 5 and 6 respectively; b and d, wild-type sequences. Arrows, position of the mutations. II. Representative of immunohistochemical staining (brown stain) of PTEN protein level in HER-2 positive breast carcinoma. PTEN level was scored as (A) -, no cytoplasmic staining (×100); (B) +1, weak staining (×100); (C) +2, moderate staining (×100); and (D) +3, strong staining (×400).
Statistical analysis was executed to assess the correlation between PTEN, exon 5 and exon 6 mutations and pathological and biological characteristics as shown in Table 3. No statistically significant correlations were observed between mutations and pathological and biological parameters. Mutations in both exons (5 and 6) were analyzed versus clinicopathological factors such as age (p = 1), menopausal status (p = 1), histology (p = 0.796), tumor grade (p = 0.424), lymph node status (p = 0.284), HER2 score (p = 0.472), estrogen receptor (p = 0.561) and progesterone receptor (p = 0.563).
Correlation between PTEN, exon 5 and 6 mutations status and clinicopathological data.
| Clinical data | PTEN all mutant | Exon 5 | Exon 6 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | wt No. (%) | mt No. (%) | p value | n | wt No. (%) | mt No. (%) | p value | n | wt No. (%) | mt No. (%) | p value | |
| Age (years) | ||||||||||||
| ≤50 | 27 | 25 (92.5) | 2 (7.5) | 1.00 | 28 | 26 (92.9) | 2 (7.1) | 0.503 | 27 | 27 (100) | 0 (0) | 0.45 |
| >50 | 21 | 20 (95.2) | 1 (4.8) | 20 | 20 (100.0) | 0 0.0) | 22 | 21 (95.5) | 1 (4.5) | |||
| Menopausal status | ||||||||||||
| Premenopausal | 36 | 34 (94.7) | 2 (5.3) | 1.00 | 36 | 34 (94.7) | 2 (5.3) | 1.00 | 36 | 36 (100) | 0 0) | 0.265 |
| Postmenopausal | 12 | 11 (91.6) | 1 (8..4) | 12 | 12 (100) | 0 (0) | 13 | 12 (92.3) | 1 (7.7) | |||
| Histology | ||||||||||||
| Ductal | 42 | 39 (92.9) | 3 (7.1) | 0.796 | 41 | 39 (95.5) | 2 (4.5) | 0.836 | 43 | 42 (97.7) | 1 (2.3) | 0.931 |
| Lobular | 2 | 2 (100) | 0 (0) | 2 | 2 (100) | 0 (0) | 2 | 2 (100) | 0 (0) | |||
| Other | 4 | 4 (100) | 0 (0) | 5 | 5 (100) | 0 (0) | 4 | 4 (100) | 0 (0) | |||
| Tumor grade | ||||||||||||
| II | 40 | 38 (95) | 2 (5) | 0.424 | 40 | 39 (95.3) | 1 (4.7) | 0.309 | 41 | 40 (97.7) | 1 (2.3) | 1.00 |
| III | 8 | 7 (87.5) | 1 (12.5) | 8 | 7 (87.5) | 1 (12.5) | 8 | 8 (100) | 0 (0) | |||
| Lymph node status | 40 | |||||||||||
| Negative | 12 | 11 (91.6) | 1 (9.4) | 0.284 | 11 | 10 (90.9) | 1 (9.1) | 0.682 | 13 | 13 (100) | 0 (0) | |
| Positive 1-3 | 7 | 7 (100) | 0 (0) | 7 | 7 (100) | 0 (0) | 7 | 7 (100) | 0 (0) | |||
| Positive > 3 | 21 | 19 (90.5) | 2 (9.5) | 22 | 21 (95.5) | 1 (4.5) | 21 | 20 (95.2) | 1 (4.8) | |||
| HER2 score | ||||||||||||
| 2 | 9 | 8 (88.9) | 1 (11.1) | 0.472 | 10 | 9 (90.0) | 1 (1.0) | 0.377 | 8 | 8 (100) | 0 (0) | 1.00 |
| 3 | 39 | 37 (94.9) | 2 (5.1) | 38 | 37 (97.4) | 1 (2.6) | 40 | 39 (97.6) | 1 (2.4) | |||
| ER | ||||||||||||
| Positive | 19 | 17 (89.5) | 2 (10.5) | 0.561 | 21 | 19 (90.5) | 2 (9.5) | 0.187 | 23 | 22 (95.7) | 1 (4.3) | 1.00 |
| Negative | 27 | 26 (96.3) | 1 (3.7) | 27 | 27 (100.0) | 0 (0.0) | 26 | 26 (100) | 0 (0) | |||
| PR | ||||||||||||
| Positive | 20 | 18 (90.0) | 2 (10.0) | 0.563 | 20 | 18 (90.0) | 2 (10.0) | 0.168 | 20 | 20 (100) | 0 (0) | 1.00 |
| Negative | 28 | 27 (96.4) | 1 (3.6) | 28 | 28 (100) | 0 (0) | 29 | 28 (96.7) | 1 (3.3) | |||
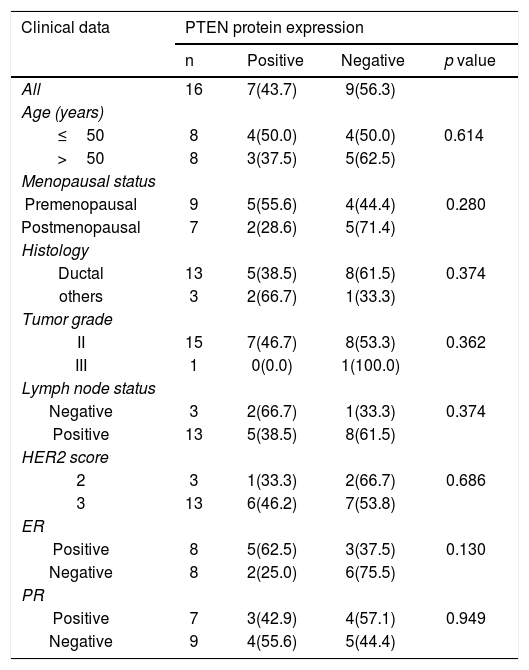
Sixteen patients were further assessed for PTEN protein expression. The immunoreactivity of PTEN was found to be ranged from complete loss or little expression to various degrees of immunostaining (Fig. 1b). PTEN loss was observed in 9 out of 16 patients (56.3%), 4 (44.4%) of which were also mutated for PI3KCA and 1(11.1%) for PTEN. None of these correlations were statistically significant. Also, statistical analysis was executed to investigate any correlation between PTEN expression and all clinicopathological data. However, there was no a statistically significant correlation (Table 4). At the histology level, PTEN loss was found mostly in ductal carcinomas (61.5%, p = 0.374). Also, it was found to be more prevalent in lymph-positive patients cases (61.5%, p = 0.374). PTEN loss was also more prevalent in patients with positive ER (75.5%, p = 0.130).
Correlation between PTEN protein expression status and clinical characteristics.
| Clinical data | PTEN protein expression | |||
|---|---|---|---|---|
| n | Positive | Negative | p value | |
| All | 16 | 7(43.7) | 9(56.3) | |
| Age (years) | ||||
| ≤50 | 8 | 4(50.0) | 4(50.0) | 0.614 |
| >50 | 8 | 3(37.5) | 5(62.5) | |
| Menopausal status | ||||
| Premenopausal | 9 | 5(55.6) | 4(44.4) | 0.280 |
| Postmenopausal | 7 | 2(28.6) | 5(71.4) | |
| Histology | ||||
| Ductal | 13 | 5(38.5) | 8(61.5) | 0.374 |
| others | 3 | 2(66.7) | 1(33.3) | |
| Tumor grade | ||||
| II | 15 | 7(46.7) | 8(53.3) | 0.362 |
| III | 1 | 0(0.0) | 1(100.0) | |
| Lymph node status | ||||
| Negative | 3 | 2(66.7) | 1(33.3) | 0.374 |
| Positive | 13 | 5(38.5) | 8(61.5) | |
| HER2 score | ||||
| 2 | 3 | 1(33.3) | 2(66.7) | 0.686 |
| 3 | 13 | 6(46.2) | 7(53.8) | |
| ER | ||||
| Positive | 8 | 5(62.5) | 3(37.5) | 0.130 |
| Negative | 8 | 2(25.0) | 6(75.5) | |
| PR | ||||
| Positive | 7 | 3(42.9) | 4(57.1) | 0.949 |
| Negative | 9 | 4(55.6) | 5(44.4) | |
HER2 protein expression is observed in approximately 20–30% of invasive breast cancer and indicating poor prognosis.1 Trastuzumab targets the extracellular domain of the HER-2 receptor. It has proved that it significantly improves time to disease progression and overall survival in patients with HER-2 over-expression and/or amplification in early-stage and metastatic tumors.2,11 Over-expression and amplification of HER2 lead to activation of PI3K and MAPK pathways, and subsequently triggering many tumor hallmarks including increasing cell proliferation, inhibiting apoptosis, angiogenesis, tumor invasion and metastasis.12PTEN is the tumor suppressor gene which inhibiting the PI3K pathway,3 which renders it as a potential predictive marker of trastuzumab treatment. Thus, the main objective of the present study is evaluate both prevalence of PTEN mutations and expression in HER2-positive breast cancer patients.
Loss of PTEN gene function was repeatedly reported to be associated with breast cancer. In PTEN-hypermorphic mouse, slight downregulation of PTEN was sufficient to develop breast cancer.13 From treatment perspective, it is associated with trastuzumab resistance in HER2-positive patients.14,15 Therefore, loss of PTEN function either by mutation(s) or downregulation could benefit as prediction marker. In this study, we thus screened mutations in exons -5, -6, and -7 in 51 HER2-positive breast cancer patients. PTEN mutations were found 3 out of 51 HER2-positive breast cancer patients with frequency of 5.9% which is in line with prevalence in several previous studies. The harbored mutations were found in exons 5 and 6, and no mutation was detected in exon 7. Association of mutation with clinical data was statistically insignificant.
Also, 16 patients out of 51 were selected to investigate the PTEN protein expression by immunostaining. In IHC analysis, PTEN loss was detected in 9 out of 16 HER2-positive BC patients with frequency of (56.3%). This results are in line with previous findings, in which the PTEN loss was detected in 35–86% of HER2-positive breast cancer patients.14,16–21 In translational medicine, loss of PTEN is a potential mechanism trastuzumab resistance.14
In a previous study, they found that loss of PTEN is significantly associated with triple-negative breast cancer patients with poor prognosis. Also, it was shown that it links to higher tumor grade and bigger tumor size, lymph node invasion and tumor recurrence.22 Among the different breast cancer subtypes, loss of PTEN expression was shown to be ore frequent in triple-negative breast cancer patients.23–25 In this study, our results showed that low expression of PTEN is more frequent in ER-negative patients compared to ER-positive (75.5 vs. 37.5%, p = 0.130). The insignificant p-value in our results could be referred to small cohort used. PTEN mutations are previously reported in breast cancer patient with frequency >5%.26–28
In brief, our data showed 5.9% of PTEN mutations (exons 5 and 6) in 51 HER2-positive breast cancer patients. Also, immunostaining analysis showed downregulation/loss of PTEN protein in 56.3% of the 16 patients. There was a tendency of have downregulation of PTEN protein in patients with ER-negative.
FundingThis work has not received any funding.
Ethical approvalSamples’ retrieval was approved by the ethics committee of the National Cancer Institute (IRB2010012036B.1).
DeclarationI confirm that I have obtained all consents required by applicable law for the publication of any personal details or images of patients, research subjects or other individuals that are used in the materials submitted to Elsevier. I have retained a written copy of all such consents and I agree to provide Elsevier with copies of the consents or evidence that such consents have been obtained if requested by Elsevier.
Authors’ contributionFatma Elwy: done most of the practical part. Zeinab Shehab El din: immunostaining. Magda M. Assem: samples’ collection, Nagwa H A Hassan: revision. Reham Helwa: supervise the whole project, writing, data interpretation, and statistics.
We thank Dr. Rafael Martinez for translating the abstract to Spanish.