A total of 268 Bacillus thuringiensis strains obtained from different sources of Argentina were analyzed to determine the diversity and distribution of the cry1, cry2, cry8, cry9 and vip3A genes encoding for lepidopteran-specific insecticidal proteins. Twin strains were excluded. Ten different profiles were detected among the 80 selected B. thuringiensis strains. Two of these profiles (cry1Aa, cry1Ac, cry1Ia, cry2Aa, cry2Ab and vip3Aa (35/80), and cry1Aa, cry1Ab, cry1Ac, cry1Ia, cry2Aa, cry2Ab and vip3Aa (25/80)) pooled 75% of the strains. The existence of this low diversity is rare, since in most of the studied collections a great diversity of insecticidal toxin gene profiles has been described. In addition, the most frequently detected profile was also most frequently derived from soil (70%), stored product dust (59%) and spider webs (50%). In contrast, the cry1Aa, cry1Ab, cry1Ac, cry1Ia, cry2Aa, cry2Ab and vip3Aa profiles were mainly detected in strains isolated from leaves (40%) and dead insect larvae (50%). Six of the identified insecticidal toxin gene profiles were discovered in strains isolated from stored product dust and leaves indicating higher diversity of profiles in these kinds of sources than in others. Some strains with high insecticidal activity against Epinotia aporema (Lepidoptera) larvae were identified, which is important to explore future microbial strategies for the control of this crop pest in the region.
Se analizaron 268 cepas de Bacillus thuringiensis obtenidas de diferentes fuentes de Argentina con el objeto de determinar la diversidad y distribución de genes cry1, cry2, cry8, cry9 y vip3A, que codifican proteínas insecticidas lepidóptero-específicas. Se excluyeron las cepas gemelas. Se detectaron solo diez perfiles diferentes entre los 80 B. thuringiensis seleccionados. Dos de estos perfiles, el cry1Aa, cry1Ac, cry1Ia, cry2Aa, cry2Ab y vip3Aa (35/80) y el cry1Aa, cry1Ab, cry1Ac, cry1Ia, cry2Aa, cry2Ab y vip3Aa (25/80), comprendieron el 75% de las cepas seleccionadas. La existencia de esta baja diversidad es una rareza, ya que en la mayor parte de las colecciones estudiadas se ha descrito una gran diversidad de perfiles de genes de toxinas insecticidas. El perfil detectado con mayor frecuencia se obtuvo principalmente de cepas procedentes de suelo (el 70% de los de esa fuente lo tenían), también fue mayoritario entre los procedentes de polvo de producto almacenado (59%) y en los que procedían de telas de araña (50%). En cambio, el perfil cry1Aa, cry1Ab, cry1Ac, cry1Ia, cry2Aa, cry2Ab y vip3Aa se detectó principalmente en las cepas aisladas de hojas (40%) y de larvas de insectos muertos (50%). Seis de los perfiles identificados fueron encontrados en cepas aisladas de polvo de producto almacenado y de hojas, lo que indica una mayor diversidad de perfiles en estas fuentes que en otras. Se identificaron algunas cepas con alta actividad insecticida contra larvas de Epinotia aporema (Lepidoptera), hallazgo importante para explorar en el futuro estrategias microbianas para el control de esta plaga en la región.
The extensive culture of crops requires the utilization of chemical insecticides to control the attack of lepidopteran pests, which may be toxic and may cause environmental hazards when used improperly. Therefore, the interest in the application of bioinsecticides based on the gram-positive entomopathogenic bacterium Bacillus thuringiensis as an option instead of chemical insecticides has contributed to establish B. thuringiensis collections all around the world, since they have a limited and particular spectrum of toxicity to a specific range of insect species37. Characterization of these collections has revealed the great variability, diversity and distribution of this bacterium in nature. B. thuringiensis is found in soil, in stored product dust, in the phylloplane of plants, and in insects and their habitats, even in spider webs4,23,30,38,42,43. Furthermore, some degree of relationship has been established between the distribution of B. thuringiensis and the type of sample, and between their distribution and the geographical or climate region of origin23,43.
The insecticidal properties of B. thuringiensis are mainly attributed to the synthesis of insecticidal crystal proteins (Cry proteins) and/or vegetative insecticidal proteins (Vip proteins), which are synthesized during sporulation or vegetative growth, respectively. More than 700 cry and 130 vip genes have been identified and classified into classes and subclasses based on the percent of pairwise amino acid identity of their corresponding proteins15. It has been established that genes within the cry1, cry2, cry9 and vip3 groups encode proteins that are toxic against lepidopteran larvae7. Recently, a cry8 gene has been included into this selected group of encoding lepidopteran active proteins1. Generally, Cry producer strains synthesize 130–140kDa proteins contained in bipyramidal crystals and also synthesize 65kDa proteins contained in smaller cuboidal crystals which have a somewhat extended toxicity spectrum, as some are also mildly toxic to mosquito larvae9. Still, some lepidopteran-active B. thuringiensis strains can produce 130kDa proteins which occur as spherical inclusions1,44. Most B. thuringiensis strains harbor complex insecticidal gene combinations5,8,27,43, such as the well-known HD-1 strain46, whereas some others can harbor a single cry gene, such as strain HD-73 strain29.
The aim of the present work was to characterize a B. thuringiensis collection from Argentina. We determined the diversity and distribution of lepidopteran-specific insecticidal toxin genes in these isolates and tried to correlate these gene profiles with the region and source of isolation. In addition, the toxicity of specific B. thuringiensis native isolates harboring at least one of these insecticidal toxin genes was analyzed through bioassays against E. aporema, a lepidopteran insect that may attack legume crops in Argentina and other regions of South America.
Materials and methodsB. thuringiensis isolates and strainsTwo hundred and sixty eight B. thuringiensis strains collected from soils, stored product dust, leaves, spider webs, and dead insect larvae from different regions of Argentina were obtained from the Instituto de Microbiologia y Zoologia Agricola – Instituto Nacional de Tecnologia Agropecuaria (IMYZA-INTA) bacterial collection. B. thuringiensis serovar kurstaki HD-1 and serovar israelensis HD-567 were kindly provided by the United States Department of Agriculture (USDA), Agricultural Research Service (Peoria, USA), and B. thuringiensis serovar morrisoni DSM2803 by the Centro de Investigacion y Estudios Avanzados (CINVESTAV, Irapuato, Mexico). Powders of spore-crystal complexes were obtained as previously described and kept at −20°C until further use38.
Characterization of crystals and their protein compositionParasporal inclusions of each isolate were primarily classified through phase-contrast microscopy. The protein composition of crystals was determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with 3% stacking gel and 10% running gel in a Bio-Rad Mini Protean 3 Cell system. Electrophoresis was carried out at 50V for 15min and 100V for 2h. Gels were stained with Coomassie Brilliant Blue. High molecular weight standard mixture (Sigma SDS-6H) was used to estimate molecular masses of crystal proteins.
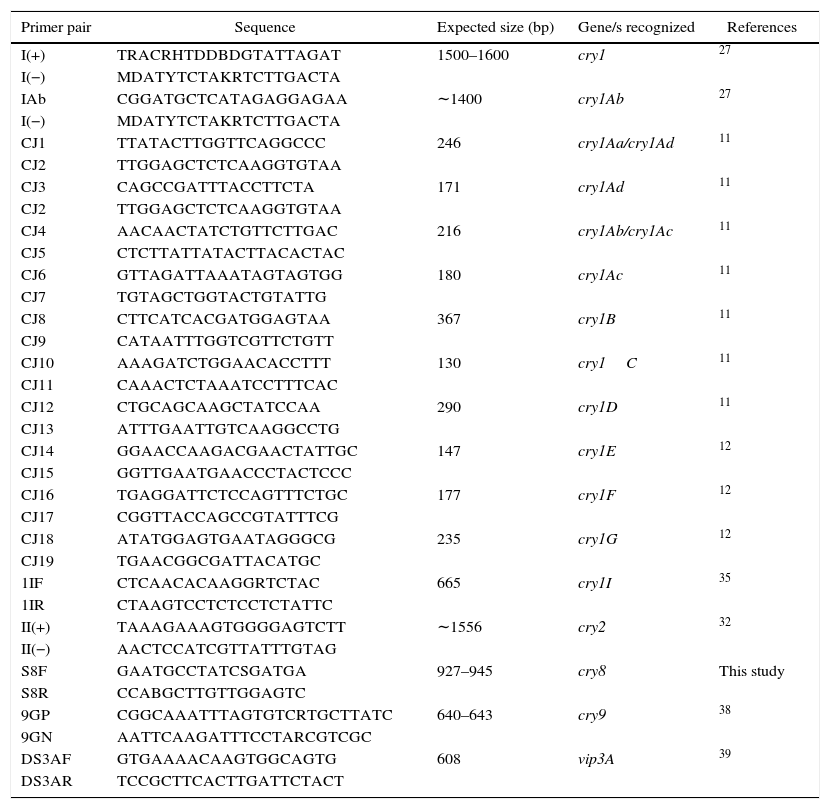
Detection and identification of insecticidal toxin genesThe DNA templates for PCR were obtained as previously described35. Five microliter of supernatant was used as DNA template in each reaction. Detection of cry1 genes was carried out by following conditions, essentially as previously described by Juarez-Perez et al.27, using the I(+) and I(−) group primers. Further identification of cry1Aa, cry1Ab, cry1Ac, cry1Ad, cry1B, cry1C, cry1D, cry1F and cry1G was conducted as described by Ceron et al.11,12 and Juarez-Perez et al.27 PCR-restriction fragment length polymorphism (PCR-RFLP) methods previously described by Sauka et al.34,35,39 were used to detect and identify cry2, cry1I and vip3A genes. Detection of cry9 genes was carried out by following conditions as previously described38, using the 9GP and 9GN group primers. For the detection of cry8 genes, novel specific primers were designed based on the analysis of conserved regions by multiple alignments of DNA sequences in the “Bt toxin nomenclature website”15 using ClustalW (http://www.ebi.ac.uk/clustalw/) and Oligoanalyzer 3.1 (http://scitools.idtdna.com/scitools/Applications/OligoAnalyzer/). This PCR was carried out according to Sauka et al.38, but using 2mM MgCl2 per reaction and each cycle consisting of an annealing step at 48°C for 1min. Primers used for the amplification of a DNA fragment of 927–945bp in size of cry8 genes and the others used during the course of this study are summarized in Table 1.
Characteristics of primers used for insecticidal toxin gene detection and identification
| Primer pair | Sequence | Expected size (bp) | Gene/s recognized | References |
|---|---|---|---|---|
| I(+) | TRACRHTDDBDGTATTAGAT | 1500–1600 | cry1 | 27 |
| I(−) | MDATYTCTAKRTCTTGACTA | |||
| IAb | CGGATGCTCATAGAGGAGAA | ∼1400 | cry1Ab | 27 |
| I(−) | MDATYTCTAKRTCTTGACTA | |||
| CJ1 | TTATACTTGGTTCAGGCCC | 246 | cry1Aa/cry1Ad | 11 |
| CJ2 | TTGGAGCTCTCAAGGTGTAA | |||
| CJ3 | CAGCCGATTTACCTTCTA | 171 | cry1Ad | 11 |
| CJ2 | TTGGAGCTCTCAAGGTGTAA | |||
| CJ4 | AACAACTATCTGTTCTTGAC | 216 | cry1Ab/cry1Ac | 11 |
| CJ5 | CTCTTATTATACTTACACTAC | |||
| CJ6 | GTTAGATTAAATAGTAGTGG | 180 | cry1Ac | 11 |
| CJ7 | TGTAGCTGGTACTGTATTG | |||
| CJ8 | CTTCATCACGATGGAGTAA | 367 | cry1B | 11 |
| CJ9 | CATAATTTGGTCGTTCTGTT | |||
| CJ10 | AAAGATCTGGAACACCTTT | 130 | cry1C | 11 |
| CJ11 | CAAACTCTAAATCCTTTCAC | |||
| CJ12 | CTGCAGCAAGCTATCCAA | 290 | cry1D | 11 |
| CJ13 | ATTTGAATTGTCAAGGCCTG | |||
| CJ14 | GGAACCAAGACGAACTATTGC | 147 | cry1E | 12 |
| CJ15 | GGTTGAATGAACCCTACTCCC | |||
| CJ16 | TGAGGATTCTCCAGTTTCTGC | 177 | cry1F | 12 |
| CJ17 | CGGTTACCAGCCGTATTTCG | |||
| CJ18 | ATATGGAGTGAATAGGGCG | 235 | cry1G | 12 |
| CJ19 | TGAACGGCGATTACATGC | |||
| 1IF | CTCAACACAAGGRTCTAC | 665 | cry1I | 35 |
| 1IR | CTAAGTCCTCTCCTCTATTC | |||
| II(+) | TAAAGAAAGTGGGGAGTCTT | ∼1556 | cry2 | 32 |
| II(−) | AACTCCATCGTTATTTGTAG | |||
| S8F | GAATGCCTATCSGATGA | 927–945 | cry8 | This study |
| S8R | CCABGCTTGTTGGAGTC | |||
| 9GP | CGGCAAATTTAGTGTCRTGCTTATC | 640–643 | cry9 | 38 |
| 9GN | AATTCAAGATTTCCTARCGTCGC | |||
| DS3AF | GTGAAAACAAGTGGCAGTG | 608 | vip3A | 39 |
| DS3AR | TCCGCTTCACTTGATTCTACT |
Crystals of selected B. thuringiensis strains were purified by continuous NaBr gradient differential centrifugation and solubilized as previously described38.
Epinotia aporema bioassaysToxicity of B. thuringiensis spore-crystal suspensions was analyzed by bioassays against neonate larvae of E. aporema Wals. (Lepidoptera: Tortricidae). Spore-crystal suspensions (final concentration 2.5μg/ml) were incorporated into polypropylene conical tubes containing a thermostatized (40°C) artificial diet for E. aporema and poured into each well of a 24-well plate (Nunc 143982)20. Only sterile distilled water was added to the negative controls and also serovar kurstaki HD-1 strain was used as positive controls. Twenty four neonate E. aporema larvae were used per assay (three replicates at least). Mortality was registered after 5 days at 29°C. E. aporema larvae were considered dead if they failed to respond to gentle probing. Bioassays with purified and solubilized crystals were conducted in the same way, except that a series of six concentrations (concentration range: 3.500–0.272μg/ml; dilution factor: 0.600) were prepared in order to establish the concentration-response relationship by probit analysis. Twenty four larvae were tested for each concentration. Statistical restrictions were followed as mentioned earlier24.
Results and discussionCharacterization of an Argentine B. thuringiensis collectionIn this manuscript, we present the characterization of a B. thuringiensis collection built from strains isolated from different sources of Argentina. This characterization contributes to a better knowledge of B. thuringiensis diversity in Argentina and South America, where only a few large collections have been extensively characterized3,6,17,41,42. Moreover, even fewer studies have reported a detailed characterization of B. thuringiensis strain collections in terms of insecticidal toxin gene content42. In particular, this Argentine B. thuringiensis collection was characterized through different methods focused only on putative lepidopteran-active isolates. PCR and PCR-RFLP results showed that 195 strains out of 268 harbors, at least one anti-lepidopteran toxin gene. Those B. thuringiensis strains isolated from the same sample that share the same morphology of parasporal crystals, the same protein composition of crystals as revealed by SDS-PAGE and the same insecticidal toxin gene profile were considered twin strains. Exclusion of twin strains is important in order to get a real estimation of the diversity of the sampled areas34. That is, 80 of these isolates were selected for further studies in order to avoid an overestimation of distribution frequencies. A full list of these isolates and their main characteristics are presented in the Supplementary Table.
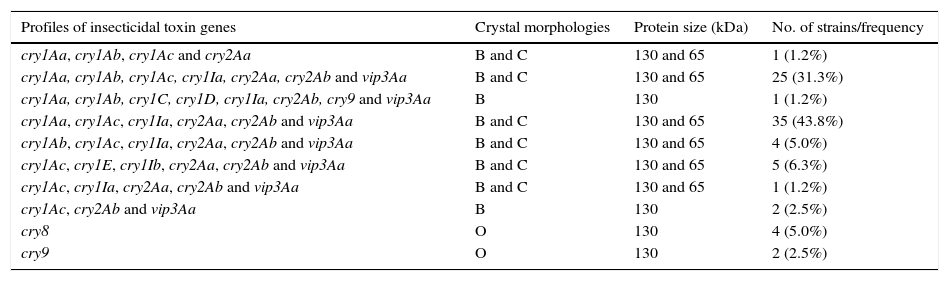
The specific characterization of these selected isolates produced ten different profiles of lepidopteran-specific insecticidal genes (Table 2). The cry1Aa, cry1Ab, cry1Ac, cry1Ia, cry2Aa, cry2Ab and vip3Aa, and cry1Aa, cry1Ac, cry1Ia, cry2Aa, cry2Ab and vip3Aa profiles were the most frequently detected. Both profiles pooled 75.0% of the selected strains. The low number of different profiles and this high frequency of strains harboring these two insecticidal toxin gene profiles that differ by a single gene would only indicate low diversity in native strains of Argentina. The existence of this low diversity is a rarity, since in most of the studied collections a great diversity of insecticidal toxin gene profiles has been described4,5,8,18,19,23,42,43,45. Some others were detected at very low frequencies. That is the case of the cry1Aa, cry1Ab, cry1C, cry1D, cry1Ia, cry2Ab, cry9 and vip3Aa profiles. The simultaneous presence of cry1C and cry1D genes is a common feature in most of the screening studies of insecticidal toxin genes but it is not in our collection5,19,23,43.
Distribution of insecticidal toxin genes in the Argentine Bacillus thuringiensis collection (n=80)
| Profiles of insecticidal toxin genes | Crystal morphologies | Protein size (kDa) | No. of strains/frequency |
|---|---|---|---|
| cry1Aa, cry1Ab, cry1Ac and cry2Aa | B and C | 130 and 65 | 1 (1.2%) |
| cry1Aa, cry1Ab, cry1Ac, cry1Ia, cry2Aa, cry2Ab and vip3Aa | B and C | 130 and 65 | 25 (31.3%) |
| cry1Aa, cry1Ab, cry1C, cry1D, cry1Ia, cry2Ab, cry9 and vip3Aa | B | 130 | 1 (1.2%) |
| cry1Aa, cry1Ac, cry1Ia, cry2Aa, cry2Ab and vip3Aa | B and C | 130 and 65 | 35 (43.8%) |
| cry1Ab, cry1Ac, cry1Ia, cry2Aa, cry2Ab and vip3Aa | B and C | 130 and 65 | 4 (5.0%) |
| cry1Ac, cry1E, cry1Ib, cry2Aa, cry2Ab and vip3Aa | B and C | 130 and 65 | 5 (6.3%) |
| cry1Ac, cry1Ia, cry2Aa, cry2Ab and vip3Aa | B and C | 130 and 65 | 1 (1.2%) |
| cry1Ac, cry2Ab and vip3Aa | B | 130 | 2 (2.5%) |
| cry8 | O | 130 | 4 (5.0%) |
| cry9 | O | 130 | 2 (2.5%) |
B: bipyramidal; C: cuboidal; O: ovoid.
The combination of insecticidal toxin genes seems to be not random. For example, strong associations were observed between the three subclasses of cry1A genes. As shown in Table 2, cry1Aa/cry1Ac, and cry1Aa/cry1Ab/cry1Ac have a strong tendency to occur together. The existence of strains harboring these cry gene combinations may be explained by previous studies on B. thuringiensis serovar kurstaki HD-1. Carlton and González10 suggested that the cry1Aa and cry1Ac genes would be located together in a smaller and unstable ∼44-MDa plasmid that can be cured easily. While cry1, cry2 and vip3Aa genes tend to be found together, these results also suggest that this association does not exist between them and cry8 or cry9 genes. Some specific classes of the cry1 and cry2 genes were found together very frequently. The presence of one cry1 gene at least with one cry2 was detected in 92.5% of the Argentine strains. This kind of association is known and has been reported previously2,5,23,31,33,42. Among cry2 genes, a strong association was observed between cry2Aa and cry2Ab; 87.5% of the strains showed to harbor these genes. This combination was also the most frequent (90.4%) in B. thuringiensis strains isolated from China28. Ben-Dov et al.5 reported that this kind of association was found in strains from Israel, Kazakhstan and Uzbekistan but less frequently (34.4%).
Wang et al.43 reported a strong association between cry1A and cry1I genes in strains isolated from China. In our study, we observed this association in 71.0% of the strains. This strong association is attributed to the relationship between the three subclasses of cry1A detected in our study and the cry1Ia gene and, between cry1Ac and cry1Ib. He et al.21 reported that B. thuringiensis subsp. chinensis CT-43 harbors the cry1Aa3, cry1Ia14, cry2Aa9, cry2Ab and vip3Aa10 genes close to each other in the largest plasmid of the strain. This observation can explain the strong association between these genes reported in 75.0% of the strains in our study. Lately, Zhu et al.46 have confirmed that strain HD-1 harbors six crystal protein genes. Four of these proteins (cry1Aa, cry1Ia, cry2Aa, and cry2Ab) are located on the large plasmid pBMB299 with the vegetative insecticidal protein gene vip3Aa where they form a pathogenicity island; two additional proteins are located on plasmids pBMB95 (cry1Ac) and pBMB65 (cry1Ab). Although it is clear that some profiles of insecticidal toxin genes are more common in nature than others, probably providing a major biologic advantage to their host, further studies are required to understand this relationships.
It is worth noting that 88.8% of the Argentine strains that harbor cry1 and cry2 genes also show bipyramidal and cuboidal crystals as observed by phase-contrast microscopy, and protein patterns of approximately 130 and 65kDa as revealed by SDS-PAGE (Table 2). These characteristics are typical of strains expressing Cry1 and Cry2 proteins40. Strains harboring cry1Aa, cry1Ab, cry1C, cry1D, cry1Ia, cry2Ab, cry9, vip3Aa (1.2%) or cry1Ac, cry2Ab, vip3Aa (1.2%) profiles produced only bipyramidal crystals and a unique band of approximately 130kDa (Table 2). However, cuboidal inclusions and a approximately 65kDa band at the SDS-PAGE gel typical of strains expressing Cry2 proteins22 were not detected in these Argentine strains. It is noteworthy that other B. thuringiensis strains that harbor a cry2Ab gene contain little or no Cry2Ab protein in their crystalline inclusions16. Lack of expression of cry2Ab genes has been related to mutations that lead to a loss in the coding frame14, to the lack of a functional promoter25, and to the lack of expression due to a post-transcriptional factor38. Since the cry1I and vip3Aa genes encode insecticidal proteins secreted during the vegetative phase of growth of B. thuringiensis37, they do not form parasporal crystal, thus their protein pattern is not expected to be in our SDS-PAGE gels. Those strains that harbor simply cry9 (2.5%) or cry8 (5.0%) genes exhibited an ovoid shape, containing a major protein of approximately 130kDa (Table 2). To our knowledge, the association among cry9 genes, this type of crystal shape and this major protein component kDa have not been reported previously. By contrast, Cry8-associated ovoid crystals resembled the parasporal bodies of some atypical Lepidopteran toxic strains previously reported by our group and other researchers1,44.
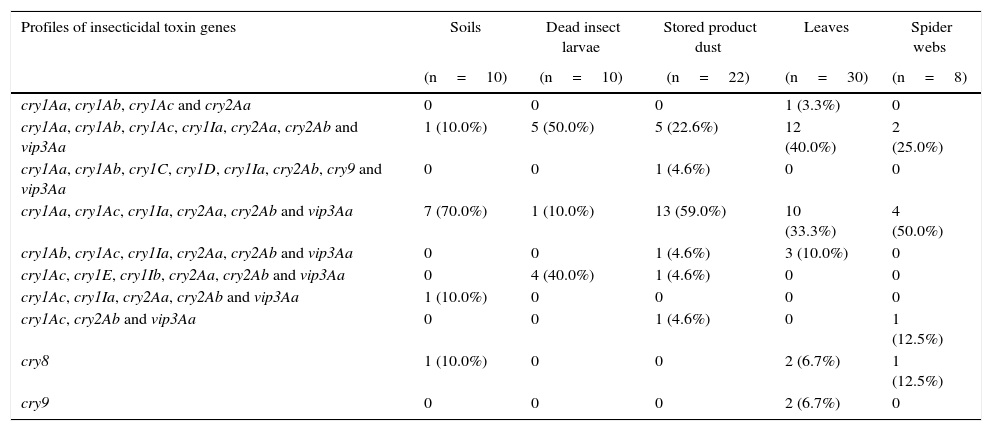
Distribution of insecticidal toxin gene profiles in B. thuringiensis strains isolated from different sourcesThe analysis of the insecticidal toxin gene distribution according to the sample source of the strains is presented in Table 3. We found that the strains containing the most common profiles cry1Aa, cry1Ac, cry1Ia, cry2Aa, cry2Ab and vip3Aa were more frequently derived from soil (70.0%), stored product dust (59.0%) and spider webs (50.0%). On the other hand, the cry1Aa, cry1Ab, cry1Ac, cry1Ia, cry2Aa, cry2Ab and vip3Aa were mainly detected in strains isolated from leaves (40.0%) and dead insect larvae (50.0%). Six of ten of the identified insecticidal toxin gene profiles were observed in strains isolated from stored product dust and leaves. These data may indicate that the diversity of insecticidal gene profiles is somewhat higher in these kinds of sample sources than in soil, dead insect larvae and spider web samples. In previous studies, some authors have suggested that stored product dust23,43 and phylloplane of plants26 are a rich source of diversity of B. thuringiensis. These results imply that stored product dust and leaf samples are a rich source of diversity of B. thuringiensis and confirms previous findings.
Insecticidal toxin gene distribution according to sample origin
| Profiles of insecticidal toxin genes | Soils | Dead insect larvae | Stored product dust | Leaves | Spider webs |
|---|---|---|---|---|---|
| (n=10) | (n=10) | (n=22) | (n=30) | (n=8) | |
| cry1Aa, cry1Ab, cry1Ac and cry2Aa | 0 | 0 | 0 | 1 (3.3%) | 0 |
| cry1Aa, cry1Ab, cry1Ac, cry1Ia, cry2Aa, cry2Ab and vip3Aa | 1 (10.0%) | 5 (50.0%) | 5 (22.6%) | 12 (40.0%) | 2 (25.0%) |
| cry1Aa, cry1Ab, cry1C, cry1D, cry1Ia, cry2Ab, cry9 and vip3Aa | 0 | 0 | 1 (4.6%) | 0 | 0 |
| cry1Aa, cry1Ac, cry1Ia, cry2Aa, cry2Ab and vip3Aa | 7 (70.0%) | 1 (10.0%) | 13 (59.0%) | 10 (33.3%) | 4 (50.0%) |
| cry1Ab, cry1Ac, cry1Ia, cry2Aa, cry2Ab and vip3Aa | 0 | 0 | 1 (4.6%) | 3 (10.0%) | 0 |
| cry1Ac, cry1E, cry1Ib, cry2Aa, cry2Ab and vip3Aa | 0 | 4 (40.0%) | 1 (4.6%) | 0 | 0 |
| cry1Ac, cry1Ia, cry2Aa, cry2Ab and vip3Aa | 1 (10.0%) | 0 | 0 | 0 | 0 |
| cry1Ac, cry2Ab and vip3Aa | 0 | 0 | 1 (4.6%) | 0 | 1 (12.5%) |
| cry8 | 1 (10.0%) | 0 | 0 | 2 (6.7%) | 1 (12.5%) |
| cry9 | 0 | 0 | 0 | 2 (6.7%) | 0 |
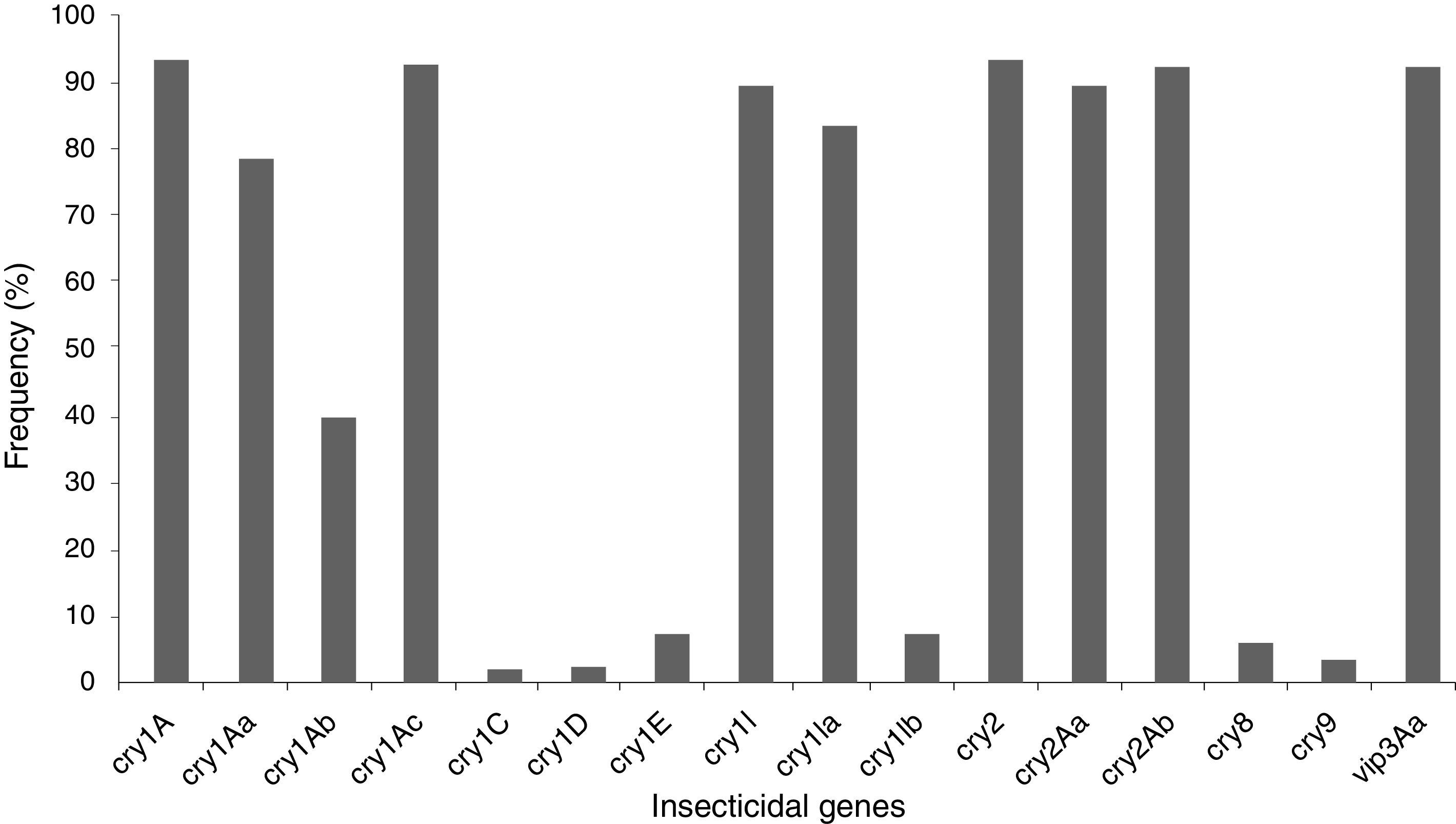
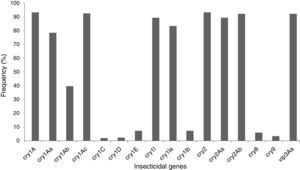
The insecticidal toxin gene composition of the B. thuringiensis strains was also analyzed (Fig. 1). Strains containing cry1A or cry2A genes were the most abundant and represent 74 of the 80 (92.5%) studied B. thuringiensis (Fig. 1); 91.3% of the strains harbor a vip3A gene and 88.8% a cry1I gene. Just 5.0% and 2.5% of the strains contained a cry8 or cry9 gene respectively. The cry1Ac (91.3%), cry2Ab (91.3%), vip3Aa (91.3%), cry2Aa (88.8%), cry1Ia (82.5%), cry1Aa (77.5%), and cry1Ab (38.8%) genes were the most frequently detected, while cry1Ib (6.3%), and cry1E (6.3%) genes were less abundant. Very few cry1C (1.3%) and cry1D (1.3%) genes were found. Not all of the searched classes and subclasses of cry1, cry2 and vip3 genes were identified in the Argentine strains.
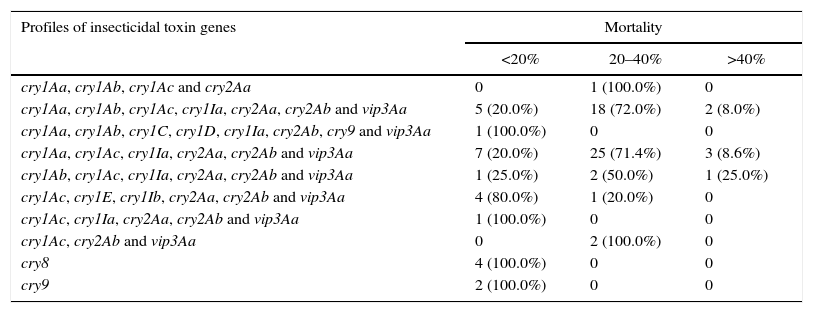
Correlation between the toxicity of Argentine B. thuringiensis strains against E. aporema and their insecticidal toxin gene profilesPreliminary bioassays with spore-crystal suspensions (final concentration 2.5μg/ml) of the 80 Argentine B. thuringiensis strains were performed with neonate larvae of E. aporema. These strains showed a mortality range of 0.0–52.5% (see Supplementary Table). A classification of the strains according to their toxicity and insecticidal toxin gene profiles is shown in Table 4. Most strains are classified in the category of 20–40% mortality. However, there were six strains that caused more than 40% mortality. The most toxic strains were isolated from phylloplane, soil, stored product dust and spider webs and harbored cry1A, cry1Ia, cry2Aa, cry2Ab and vip3Aa genes. On the contrary, the six less toxic/nontoxic strains showed mortality rates under 4% and were mainly isolated from phylloplane and one from soil. These strains harbored cry8 or cry9 genes. These results suggest that multiple insecticidal toxin gene profiles may be used as markers for the spectrum of insecticidal activity of B. thuringiensis strains as previously suggested13. In addition, the mortality rate was not always associated with the insecticidal toxin gene content of particular strains. Strains harboring the same insecticidal gene profile differed in their level of toxicity against E. aporema (Table 4). For example, strains INTA H3-5 and INTA H6-3 that shared the same insecticidal toxin gene profile (cry1Aa, cry1Ab, cry1Ac, cry1Ia, cry2Aa, cry2Ab and vip3Aa) showed 38.3% and 12.7% mortality. This kind of results may be observed in previous reports23,31; however, the reasons of those variations still remain to be studied. Furthermore, it is clear that the toxic levels of B. thuringiensis in the target insects are influenced by many factors including the insecticidal toxin gene profiles, the level of expression of these genes, possible synergism between insecticidal proteins, and other unknown or undetected virulence factors. These reasons may hamper the PCR ability as a precise predictive tool for insecticidal activity in B. thuringiensis strains. The genome sequencing of all strains may overcome some of these factors. However, this partial solution is difficult for now because of the high cost of sequencing a large number of strains. Despite of all this, our results showed that different B. thuringiensis strains obtained from the same sample showed certain relationship between the level of toxicity and specific insecticidal toxin gene profiles. In general, strains harboring genes cry1Aa, cry1Ab, cry1Ac, cry1Ia, cry2Aa, cry2Ab and vip3Aa were more toxic than those harboring genes cry1Ab, cry1Ac, cry1Ia, cry2Aa, cry2Ab and vip3Aa, and these more than those harboring genes cry1Aa, cry1Ac, cry1Ia, cry2Aa, cry2Ab and vip3Aa. In a previous study, Cry1Ab and Cry1Ac had shown to be the most active proteins against neonate larvae of E. aporema, followed by Cry1Aa with LC50 values of 0.55, 1.39 and 4.14μg/ml, respectively36.
Classification of the selected B. thuringiensis strains into groups according to their mortality rate against E. aporema
| Profiles of insecticidal toxin genes | Mortality | ||
|---|---|---|---|
| <20% | 20–40% | >40% | |
| cry1Aa, cry1Ab, cry1Ac and cry2Aa | 0 | 1 (100.0%) | 0 |
| cry1Aa, cry1Ab, cry1Ac, cry1Ia, cry2Aa, cry2Ab and vip3Aa | 5 (20.0%) | 18 (72.0%) | 2 (8.0%) |
| cry1Aa, cry1Ab, cry1C, cry1D, cry1Ia, cry2Ab, cry9 and vip3Aa | 1 (100.0%) | 0 | 0 |
| cry1Aa, cry1Ac, cry1Ia, cry2Aa, cry2Ab and vip3Aa | 7 (20.0%) | 25 (71.4%) | 3 (8.6%) |
| cry1Ab, cry1Ac, cry1Ia, cry2Aa, cry2Ab and vip3Aa | 1 (25.0%) | 2 (50.0%) | 1 (25.0%) |
| cry1Ac, cry1E, cry1Ib, cry2Aa, cry2Ab and vip3Aa | 4 (80.0%) | 1 (20.0%) | 0 |
| cry1Ac, cry1Ia, cry2Aa, cry2Ab and vip3Aa | 1 (100.0%) | 0 | 0 |
| cry1Ac, cry2Ab and vip3Aa | 0 | 2 (100.0%) | 0 |
| cry8 | 4 (100.0%) | 0 | 0 |
| cry9 | 2 (100.0%) | 0 | 0 |
Samples tested n=80.
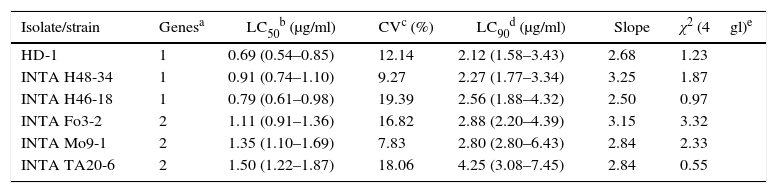
The five most toxic isolates were further bioassayed with serial dilutions of solubilized crystals for LC50 estimation (Table 5). According to the LC50 values and their fiducial limits, B. thuringiensis serovar kurstaki HD-1, B. thuringiensis INTA H46-18 and INTA H48-34 were the most toxic against E. aporema, although the difference among them was not statistically significant. The two Argentine strains were isolated from samples of phylloplane and both shared the same insecticidal toxin gene profile. In contrast, the other less toxic strains harbored a similar insecticidal toxin gene profile lacking the cry1Ab gene. B. thuringiensis H46-18 and INTA H48-34 could be used as active ingredients of pesticides formulated for controlling E. aporema.
Probit analysis of the most toxic strains against E. aporema
| Isolate/strain | Genesa | LC50b (μg/ml) | CVc (%) | LC90d (μg/ml) | Slope | χ2 (4gl)e |
|---|---|---|---|---|---|---|
| HD-1 | 1 | 0.69 (0.54–0.85) | 12.14 | 2.12 (1.58–3.43) | 2.68 | 1.23 |
| INTA H48-34 | 1 | 0.91 (0.74–1.10) | 9.27 | 2.27 (1.77–3.34) | 3.25 | 1.87 |
| INTA H46-18 | 1 | 0.79 (0.61–0.98) | 19.39 | 2.56 (1.88–4.32) | 2.50 | 0.97 |
| INTA Fo3-2 | 2 | 1.11 (0.91–1.36) | 16.82 | 2.88 (2.20–4.39) | 3.15 | 3.32 |
| INTA Mo9-1 | 2 | 1.35 (1.10–1.69) | 7.83 | 2.80 (2.80–6.43) | 2.84 | 2.33 |
| INTA TA20-6 | 2 | 1.50 (1.22–1.87) | 18.06 | 4.25 (3.08–7.45) | 2.84 | 0.55 |
The authors declare that no experiments were performed on humans or animals for this study.
Confidentiality of dataThe authors declare that no patient data appear in this article.
Right to privacy and informed consentThe authors declare that no patient data appear in this article.
Conflict of interestThe authors declare that they have no conflicts of interest.
DS holds a research career award from Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET).