Background. Patients exposed to hepatitis C virus (HCV) may develop chronic infection with viremia. The diagnosis of this condition requires the use of several laboratory tests in algorithms tailored to the population and resources available for each laboratory.
Aim. We compared the diagnostic efficacy of two diagnostic algorithms for the identification of viremic patients with HCV. One based on confirmation of reactive antibody results with molecular techniques (reverse transcription polymerase chain reaction, RT-PCR) and the other based on the use of a new HCV core antigen test (HCV Ag).
Material and methods. We measured levels of anti-HCV, HCV Ag and viral load (trough RT-PCR) in parallel, in 211 samples (53 antibody positive, 158 antibody negative). Using the three results available for each sample we simulated the diagnostic performance of the two algorithms and compared them to the results of RT-PCR as gold standard.
Results. Both algorithms showed a high degree of concordance for viremic patients. The percentage of correctly classified patients was 99.05% for the algorithm based on RT-PCR and 98.10% for the HCV Ag algorithm. The HCV core Ag test showed a clinical sensitivity of 0.917 and showed a good correlation to the results of molecular biology. Spearman rank correlation coefficient (ρ) of 0.97 (95% CI 0.95 to 0.99, p < 0.0001). Conclusion. An algorithm incorporating HCV Ag as confirmatory test for anti-HCV results is a feasible alternative to the use of molecular techniques in laboratories that do not have access to them or require faster turn around times.
It is estimated that more than 170 million people worldwide are chronic carriers of the hepatitis C virus (HCV).1 In Mexico the prevalence is close to 1% with an estimated 982,000 infected patients.2 Detection of HCV antibody (anti-HCV) has been the standard method of screening for HCV infection since its discovery more than two decades ago.3,4 The detection of HCV-Ab with modern serological test is both highly sensitive and specific5 but is limited in some settings such as “window period” infections and immunecompromised patients as those undergoing hemodialysis, where false negatives may occur.4 False positive results are also rare but can potentially occur and thus all positive results should be adequately confirmed.5 Viremia in these patients can be detected through the use of molecular techniques such as reverse transcription polymerase chain reaction (RT-PCR) but these techniques require expert staff, specialized equipment and reagents that might not be available in all settings.6 To overcome these problems, a new diagnostic test based on the detection of the HCV core antigen (HCV Ag) has been recently introduced. This test has a high specificity an analytical sensitivity of 3 fmol/L, a good relationship to molecular techniques and runs on the same instrument as the anti-HCV.7
HCV diagnostic tests should not be used in isolation and in order to help clinicians and laboratory personnel to make the best use of them, several algorithms have been developed to serve as guidance in the proper order and type of test order, depending on the objective and type of population of the institution.8,9 The objective of this study was to compare in parallel the diagnostic performance of two different algorithms for the detection of HCV viremic patients using RT-PCR or HCV Ag. A secondary objective was to analyze the diagnostic characteristics of the HCV Ag reagent in our institution.
Material and MethodsThis study was performed at the Clínica Especializada Condesa in Mexico City. This clinic provides free services of early diagnosis, counseling and treatment for HIV and other sexually transmitted diseases. Its activities are funded by the government of Mexico City.
For this study we used anonymous surplus serum samples obtained as part of the diagnostic studies performed on patients of the clinic. Some of these samples were refrigerated for a few days while they were frozen at −70 oC until processing, when they were thawed and measured in parallel with the following tests:
- •
Anti-HCV. Chemiluminescent microparticle immunoassay (CMIA) for the qualitative detection of antibody to hepatitis C virus (anti-HCV) in human serum and plasma. We used ARCHITECT Anti-HCV (Abbott Diagnostics, Chicago, IL). Test performed on serum on an ARCHITECT i2000 instrument. Cut off value for reactive results: 1.00 signal to cut off ratio.
- •
HCV core Ag. Chemiluminescent Microparticle Immunoassay (CMIA) for the quantitative determination of core antigen to hepatitis C virus in human serum and plasma of HCV core viral antigen. We used ARCHITECT HCV Ag (Abbott Diagnostics, Chicago, IL). Test performed on serum on an ARCHITECT i2000 instrument. Cut off value for positive result > 0.06 pg/mL.
- •
Reverse transcription PCR. Molecular test to detect the presence of HCV nucleic acid by reverse transcription PCR. Performed on an Abbott m2000 (Abbott Diagnostics, Chicago, IL). Test performed on serum. Sensitivity: 30 IU/mL for the 0.2 mL sample volume. Linear range: 12 IU/mL (1.08 log IU/mL) to 100 million IU/mL (log 8.0 IU/mL), standardized to second WHO international standard for HCV RNA (NIBSC 96/798). Designed to achieve an inter-assay standard deviation (SD) of ≤ 0.25 log IU/mL of HCV RNA for samples containing HCV concentrations from 100 to 100 million IU/mL.
We randomly selected for this study the first 53 samples positive for anti HCV antibody and another 158 samples negative to antibody from the routine samples obtained from patients at the clinic. RT-PCR was considered as our gold standard method to compare the results of both HCV Ag and anti-HCV. The protocol was approved by the ethics committee and patients provided consent for sample withdraws.
Data analysis was performed with Analyze it software version 2.27.
Diagnostic algorithmsWe tested two different diagnostic algorithms. The first one consisted of screening by anti-HCV followed by viral load for all positive results. Although some institutions like the CDC9 still recommend the use of confirmatory test for antibody results, many other organizations like the American Association for the Study of Liver Disease (AASLD) and the Cleveland Clinic consider the use of antibody confirmatory test as obsolete due to the high sensitivities and specificities of modern serological test and have published algorithms based only on anti-HCV screening followed by molecular tests.5,8
In line with this recommendation we developed an algorithm were the first screening step consisted on anti-HCV determination and all positive results were submitted to RT-PCR viral load measurement.
For the other algorithm we chose to use the new HCV Ag test that measures in a quantitative format the amount of core protein present in a sample. As mentioned in the literature,6,7 one of the potential uses of this test is the confirmation of anti-HCV reactive results. This algorithm has the advantage that both anti HCV and HCV Ag tests are run on the same automated platform commonly found in many laboratories so there would be no need to implement molecular methods. There is also a significant advantage in terms of time as the HCV Ag test does not require extraction and thus has a much faster turn around time.
Since we performed viral load measurements with RT-PCR on all samples, we could directly compare the results of both algorithms with the results of our gold standard in order to detect patients on “window” period of the infection that would have been missed by the anti-HCV alone.
Economic analysisWe performed a regular cost effectiveness evaluation of the two algorithms plus a third involving viral load measurement for all HCV Ag positive samples as a comparison a regular practice. A decision tree was built using TreePlan add-in for Microsoft Excel. Costs included were direct costs of testing for all tests used in the different algorithms and these were directly measured from the recorded data at the institution. Effectiveness data used was the number of correctly diagnosed patients and the incremental cost effectiveness ratios were calculated for all strategies taking the least expensive one as base. Sensibility analysis was performed to assess robustness of results in conditions of high and low disease prevalence.
ResultsOverallWe were able to process all 211 samples with the three study methods. Only 36 of the 53 HCV antibody positive patients had detectable viremia at the time of our study (67.9%). The viral load was not normally distributed with a median of 589.4 x 103 lU/mL (range < 30 to 6,161 x 103 lU/mL). Among the 158 antibody negative patients we found two cases positive for PCR, one with a viral load of 282 lU/mL and the other with < 30 lU/mL. The HCV Ag measurements did not have a normal distribution either with a median concentration of 33.7 pg/mL (range 0.125 to 303.65 pg/mL). The average ratio between viral load and HCV Ag measurements was 14,021 UI/pg HCV Ag but this parameter showed a wide variance among samples.
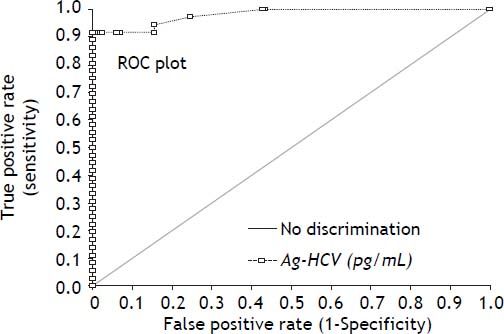
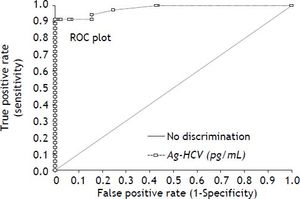
HCV Ag reagentUsing the RT-PCR results as gold standard, we obtained an overall sensitivity of 0.917 with a specificity of 0.989 at the insert cut off of 0.06 pg/mL for the new HCV Ag reagent in this high risk setting. The receiver operating characteristic (ROC) curve analysis (Figure 1) for the HCV Ag reagent showed an area under the curve of 0.98 and suggests that the optimal cut off point on our population would be 2.5 pg/mL. Use of this cut off would have yielded a specificity of 1.0 with a sensitivity of 0.917 (sentence deleted).
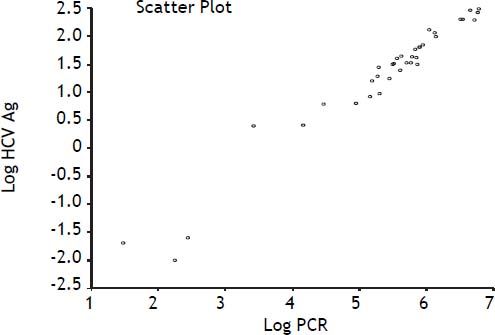
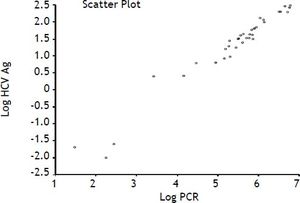
Our data show a strong correlation between the results of RT-PCR and the measurements of HCV Ag as evidenced by a Spearman rank correlation coefficient (ρ) of 0.97 (95% CI 0.95 to 0.99, p < 0.0001) (Figure 2).
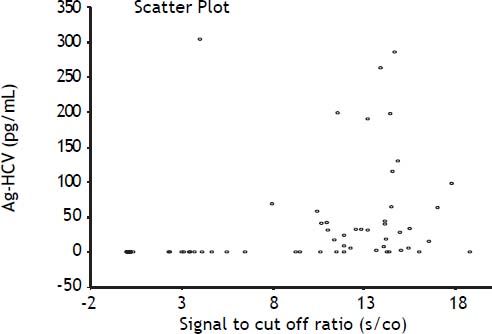
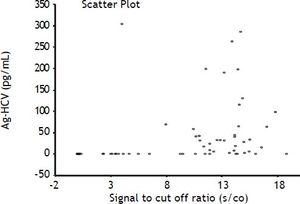
Anti-HCVIn our data we did not find any correlation between the semi quantitative results of the antibody tests (expressed as the signal to cut off ratio, s/co) and the results of both the HCV Ag and RT-PCR as evidenced by Spearman correlation coefficients of 0.51 and 0.59, respectively (Figure 3). ROC curve analysis of anti-HCV results suggested the optimal cut off level in our population corresponds to a s/co ratio ≥ 4, which would reduce the number of false positive results from 19 to 12, but it is important to emphasize that in our population even at the insert cut off level (≥ 1) most of the positive results (64%) were true positives.
AlgorithmsThe fact that we measured all of our samples with three different methodologies allowed us to simulate the results that would have been obtained by using the diagnostic algorithms presented on the Methods section. Table 1 summarizes the results obtained by each algorithm compared to the method used as gold standard for classification of patients. Using RT-PCR as the gold standard for the detection of viremic patients, the two algorithms presented would have correctly classified as viremic 99.05% of patients (Anti-HCV + RT-PCR) and 98.10% of patients (Anti-HCV + HCV Ag).
Both algorithms would have missed two patients at screening because in our data set we found two patients with very low viremia (< 300 IU/mL) that were anti-HCV negative. The confirmation using HCV Ag generates an additional false positive patient that has positive results on HCV Ag but is not viremic and an additional patient that would be classified as non viremic due to negative results of the HCV Ag but was actually positive for RT-PCR. The three viremic patients that had negative results with the HCV Ag test showed very low levels of viremia (282, 178 and < 30 IU/mL). One of these patients had high titers of anti-HCV and the other two were also antibody negative and probably represented very early infections.
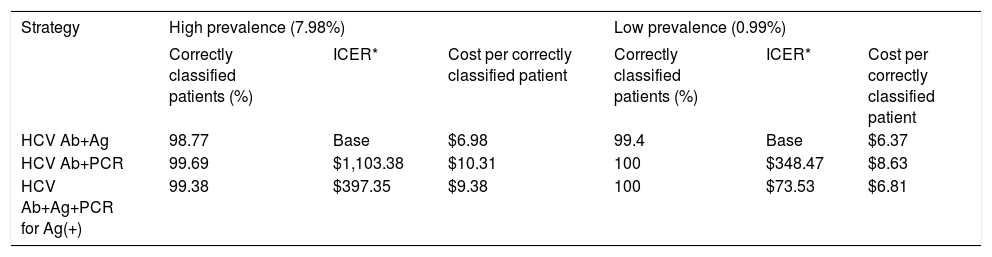
Economic analysisAn economic analysis of both algorithms plus a third algorithm involving PCR confirmation of all HCV Ag positive results was performed (as would be expected if patients were to be treated) using the perspective of the laboratory with the assumptions presented on table 2. Sensitivity analysis was performed using a decision tree with both high and low prevalence settings. Incremental cost effectiveness ratios (ICER) defined as cost per additional patient correctly classified and cost per correctly classified patients were calculated based on the results of these simulations (Table 3).
Assumptions for economic analysis.
| Perspective: laboratory. |
| Exchange rate peso/USD: 12.8551 to 1. |
| Cost included: direct costs of labor (measured) and reagents (reagents, calibrators, controls). Costs of equipment ownership not included. |
| Discounting: not used as all costs were measured for the same period. |
| Time horizon: immediate due to the transversal design of the study. |
| Sensitivity analysis: disease incidence. |
Economic analysis data for the two algorithms plus an additional algorithm with confirmatory testing with PCR for all positives.
| Strategy | High prevalence (7.98%) | Low prevalence (0.99%) | ||||
|---|---|---|---|---|---|---|
| Correctly classified patients (%) | ICER* | Cost per correctly classified patient | Correctly classified patients (%) | ICER* | Cost per correctly classified patient | |
| HCV Ab+Ag | 98.77 | Base | $6.98 | 99.4 | Base | $6.37 |
| HCV Ab+PCR | 99.69 | $1,103.38 | $10.31 | 100 | $348.47 | $8.63 |
| HCV Ab+Ag+PCR for Ag(+) | 99.38 | $397.35 | $9.38 | 100 | $73.53 | $6.81 |
The economic analysis showed that HCV Ab + RT-PCR is the most effective algorithm in terms of the clinical outcome, but this is also the most expensive both in cost per patient correctly identified and in net cost. On the other hand, the HCV Ab + HCV Ag algorithm was the least expensive option but mis-classified a small percentage of patients. The ICER shows that every additional patient correctly classified by the HCV Ab + RT-PCR algorithm would cost between $348,47-$1.103,38 depending on the prevalence. This is caused by the effect of a small improvement in effectiveness gained at the expense of a significant increase in net costs. The addition of RT-PCR to all HCV Ag positive results brings the economic parameters closer to those of the HCV Ab + RT-PCR algorithm, but even with this approach the HCV Ab + RT-PCR strategy is dominated in economic terms by the HCV Ag strategy. Interestingly, one of the most important cost drivers of the whole algorithm lies in the screening test used in the first step, as all positive results have to go to the reflex test whether they are true or false positives. Sensitivity analysis performed to test the impact of prevalence on the economic model showed that the algorithm based on HCV Ag becomes more efficient as the incidence of HCV viremia increases because of the larger number of reflex tests involved increases the cost geometrically while the reduction in the HCV Ab + HCV Ag performance is relatively minor.
DiscussionThe two diagnostic algorithms that we compared showed a high concordance between them and with the results of RT-PCR. It is important to bear in mind that these algorithms were developed to detect viremic patients and may be optimal for diagnostic laboratories that are interested in treating these patients. Laboratories with very low prevalence of HCV infection may prefer to use different algorithms. We have shown the potential utility of the new HCV Ag test as a reflex test for HCV-Ab positive results. As shown by our data the use of HCV core Ag test in this setting would yield very similar results to those obtained by the combination of HCV-Ab screening and molecular biology with a significantly shorter turn around time and lower cost per patient correctly identified. Therefore, this algorithm may be an interesting alternative for laboratories that lack a dedicated area for molecular biology testing or have limited resources available for screening and where it may be important to quickly sort out all HCV Ab positive patients without viremia. Our results confirm the diagnostic performance in terms of clinical specificity and sensitivity of the new HCV core antigen test that had been previously reported.6,7
The fact that we were able to test all of our samples with three different methodologies made it possible for us to detect both false positive and negative results from HCV Ag and anti-HCV. These results show that the proper classification of a patient with HCV infection requires the careful use of at least a couple of different techniques combined in a diagnostic algorithm tailored to the population and needs of every laboratory. The decision of the algorithm and test to use depends both on technical factors of the test such as limit of detection, sensibility and specificity as well as on the availability of resources, turn around time requirements and economic factors.
Our results do not show a relationship between the anti-HCV level measured as the signal to cut off ratio (s/co) and viremia that has been reported previously by the CDC report9 and other papers.10 There may be several explanations for this discrepancy. Viral RNA could have been degraded over time in our samples while they were refrigerated prior to freezing.11 However, this is unlikely as the discrepancy was also observed with HCV core Ag, which has been shown to remain stable over time.11 It is possible that this difference may have been caused by the difference among the populations studied (i.e. blood donors vs. high risk patients). We were therefore unable to establish a s/co ratio that clearly separated the viremic and non viremic patients as we had patients with high viremia and low s/co ratios. The ROC curve analysis shows that we could improve the sensitivity of both anti-HCV and HCV Ag in our population by using a higher cut off ratio and that this would not have much impact on the specificity. The optimal sensitivity and specificity for identifying viremic patients based on HCV Ab testing would have been reached using a cut off of 4 s/co in our data set. This shows the importance of measuring the diagnostic properties of the test on the population of interest to each laboratory.
By processing all samples with the three tests we were able to find three patients with low level viremia that were either negative to HCV Ag (one case) or to both anti-HCV and HCV Ag (two cases). One of the limitations of our study is that we did not have enough sample volume to use a second molecular technique on these samples and thus we can not distinguish if they are patients with very low levels of viremia and absence of anti-HCV or they are false positives of our molecular method. Likewise, we could not obtain serial samples and thus may have missed a patient with intermittent viremia, although the limit of the detection of the RT-PCR method used (12 UI/ mL) makes that possibility unlikely. These shortcomings underscore the fact that a correct diagnosis of HCV status is not always possible based on a single sample, even when highly sensitive methods are used. Another limitation of our study is that we were not able to determine the genotype of the infected patients and the behavior of diagnostic tests may differ according to the viral genotype. Also, due to the rather high levels of viremia in the samples studied we could not detect the positivity threshold for HCV Ag test.
ConclusionsWe have shown that a diagnostic algorithm that incorporates the new HCV core Ag test is a feasible alternative to the use of molecular techniques. This may be a valuable alternative for diagnostic laboratories were TAT is important or were resource availability limits the adoption of molecular techniques.
Abbreviations- •
Anti HCV: antibodies anti-HCV.
- •
HCV: hepatitis C virus.
- •
HCV Ag: HCV core antigen.
- •
RT-PCR: reverse transcription polymerase chain reaction.
- •
TAT: turnaround time.
Abbott laboratories provided all the reagents used on the serological and molecular biology test of this study.
Conflicts of InterestMiguel A. Reyes is an employee of the R&D division of Abbott Diagnostics.
Luis Juarez, Patricia Iracheta, Yasmin Medina, Verónica Ruiz: nothing to declare.