Matrix assisted laser desorption/ionization-time of flight (MALDI-TOF) mass spectrometry is widely established as a technique in clinical microbiology laboratories for the identification of microorganisms. Using this technique, it is also possible to obtain the identification of microorganisms from untreated urine samples.
MethodsIn this study, a differential centrifugation protocol and a criterion for validation of the results in order to achieve microbial identification from untreated urine samples are proposed. Additionally, the sensitivity of the analytical procedure in monobacterial urine samples has been evaluated.
ResultsA 90% sensitivity (confidence interval of 81.96%–94.84%) was obtained in urine samples with bacterial counts of ≥1×105CFU/ml, and it was possible to improve the percentages of direct identifications from urine samples with bacterial counts of <1×105CFU/ml.
ConclusionIt is concluded that the MALDI-TOF system is both fast and reliable in the identification of individual microorganisms from untreated urine samples with counts of ≥1×105CFU/ml.
El uso de la espectrometría de masas Matrix Assisted Laser Desorption/Ionization-Time of Flight (MALDI-TOF) es una técnica implantada en los laboratorios de Microbiología Clínica que permite llevar a cabo la identificación de microorganismos. Una de las aplicaciones de esta técnica es la identificación a partir de muestra directa de orina.
MétodosEn este estudio se propone un protocolo de centrifugación diferencial y un criterio de validación de los resultados para alcanzar la identificación microbiana a partir de muestra directa de orina. Adicionalmente se estudia la sensibilidad del procedimiento analítico en muestras de orina monomicrobianas.
ResultadosLas orinas con recuentos ≥1×105UFC/ml mostraron un 90% de sensibilidad (Intervalo de Confianza de 81.96%–94.84%) y se logró mejorar los porcentajes de identificación directa a partir de orinas con recuentas bacterianos <1×105UFC/ml.
ConclusionesLa espectrometría de masas MALDI-TOF es un sistema rápido y fiable para la identificación de microorganismos a partir de orinas monomicrobianas con recuentos ≥1×105UFC/ml.
Urinary tract infections (UTIs) can manifest in a wide clinical range from bacteriuria with limited clinical symptoms to sepsis, severe sepsis or septic shock1. In 20–30% of all septic patients the infection focus is localized in the urogenital tract2 and an adequate initial antibiotic therapy is essential since it ensures improved outcome3. Moreover, inappropriate antimicrobial therapy in severe UTI is linked to a higher mortality rate4. For election of the empiric treatment in these cases, identification directly from urine samples of bacteria causing infection can be useful.
Matrix assisted laser desorption/ionization-time of flight (MALDI-TOF) technology is a technique which allows microbial identification in clinical microbiology laboratories based on microbial protein profiles5. A promising application of the MALDI-TOF technique consists of carrying out identification from direct urine samples, without the need to cultivate them. In this way, a result at microbiological species level can be obtained in less than one hour.
Validation criteria of the results indicated by the manufacturers are restrictive, such that only high scoring identification is accepted. On the other hand, when microbial identification is carried out by MALDI-TOF from direct samples, the identification scores obtained are lower than scores obtained from culture, as a result of possible analytical interferences, which were not eliminated. So, in order to accept a higher rate of identification, the required level of validation criterion from the manufacturer should be lowered. In fact, various criteria have been published for validation of results that provided different rates of acceptable direct identifications6–9.
The objectives of this study were (i) to develop a differential centrifugation protocol which allows the identification of microorganisms by MALDI-TOF from direct urine samples, (ii) to propose a new validating criterion for the results that allows to increase the number of acceptable direct identifications, and (iii) to examine the influence of bacterial concentration in urine on the accuracy of direct identification by MALDI-TOF.
Material and methodsDetermination of sample sizeAssuming that the population was made up of positive urine samples in the period of one year (3272 in 2010 at Microbiology Service at the University Clinic Hospital of Valladolid, Spain) and accepting an alpha risk of 0.95 giving a precision of ±0.1 units in bilateral contrast to a proportion estimated at 0.5, for the validation of the differential centrifugation protocol, the study group would need to be made up of 95 monobacterial positive urine samples with counts of ≥1×105CFU/ml. A hundred samples were randomly processed to facilitate calculation. The samples were obtained through midstream clean catch and only monobacterial samples were selected because more than 95% UTIs are caused by single bacterial species10.
In order to study the relationship between bacterial concentration in urine samples and the accuracy of identification by MALDI-TOF from direct samples, 37 urines with counts of <1×105CFU/ml were processed (14 samples with counts between 5×104and 1×105CFU/ml, 17 samples with counts between 1×104 and 5×104CFU/ml, and six samples with counts <1×104CFU/ml). Additionally, four samples with yeast were processed. Briefly, a total of 141 monomicrobial positive urine samples were processed.
Working procedureIn order to obtain direct identification, the urine samples were processed as follows: samples were homogenized by agitation, and 5ml was centrifuged for 5min at 700rpm. The supernatant obtained was centrifuged for 15min at 6000rpm. The sediment was resuspended in 1ml of HPLC quality distilled water and was centrifuged again for 15min at 6000rpm. The proteins of the sediment were extracted using the ethanol/formic acid technique. In order to carry out this extraction the sample was mixed with 300μl of HPLC quality distilled water and 900μl of absolute ethanol, and vortexed until a homogeneous suspension was achieved. The mixture was centrifuged for 2min at 13,000rpm and the supernatant was eliminated. Finally, 25μl of 70% formic acid and 25μl of acetonitrile were added, vortexed and centrifuged for 1min at 13,000rpm. Using the supernatant obtained, four 1μl spots were dropped on four different areas of the metal sample plate (MSP 96 target polished steel; Bruker, Bremen, Germany). They were allowed to dry and 1μl of the matrix solution (saturated solution of cyano-4-hydroxy cinnamic acid in 50% acetonitrile with 2.5% trifluoroacetic acid) was added to each spot to be analyzed. They were allowed to dry again and then, a reading of the four spots from each sample was taken. All chemicals were supplied by Sigma–Aldrich (St Louis, MO, USA).
Furthermore, at the same time, urine samples were plated, using a calibrated loop so as to determine the number or CFU/ml present in the urine, on Columbia agar plates supplemented with 5% sheep's blood (bioMérieux, Marcy l’Etoile, France) and MacConkey medium (bioMérieux, Marcy l’Etoile, France); in addition a Sabouraud+Chloramphenicol agar plate (Difco, Maryland, USA) was added in those cases where fungal analysis was requested. The plates were incubated at 37°C/24h in aerobic atmosphere and identification was done by MALDI technique. Samples showing growth of two or more microorganisms were not included in the study.
Protein analysis was carried out using a mass spectrometer MALDI Microflex LT (BrukerDaltoniK GmbH, Bremen, Germany) with FLexControl v. 3.0 software (Bruker DaltoniK GmbH, Bremen, Germany). The spectrum was obtained in linear mode (N2 laser frequency and wavelength: 60Hz and 337nm, voltage of the ion source I: 20kV, voltage of the ion source II: 16.7kV; detector mass range: 2000–20,137Da) and 240 shots from different positions of the metal sample plate were recorded. Once the association is completed, the Biotyper v.3.0 software (Bruker DaltoniK GmbH, Bremen, Germany) suggests a list of ten most likely microorganism identifications, each with a score that indicates its accuracy11,12. Adopting the manufacturer's criteria, valid identifications may be deemed as those in which a score of ≥2.000 is obtained for the first microorganism on the list. The software also offered a consistency index, which provides information concerning the quality of the identification that does not affect the validity of the results. This consistency index depends on the concordance of the microbial identification obtained on the ten microorganisms on the list.
Results of the identification were interpreted in accordance with previously published criteria6–9; in those cases in which the criteria of the different authors were based on a single reading, the first of the four replicates carried out was chosen and the remaining three replicates were rejected. For this, urine samples were divided into two subgroups, one including the study group of 100 monobacterial samples with counts of ≥1×105CFU/ml, and another including the 37 urine samples with counts of <1×105CFU/ml. Moreover, for each urine sample, MALDI-TOF scores from direct sample and from colonies grown on plates were compared and, taking the identification obtained by MALDI-TOF from culture using the criteria of the manufacturer as the gold standard, a validation criterion for direct identification was defined.
In order to compare the identification results obtained using the different validation criteria, the McNemar test was used. In order to compare the frequency distribution in the population of the study group, the Chi-squared test was used. All these calculations were carried out using the SPSS software v. 15.0.
ResultsNo statistically significant difference was observed (p>0.05) when comparing isolates obtained from the study group with those from the University Clinic Hospital of Valladolid (Spain) during 2010 (Table 1). Moreover, the four species most frequently isolated in the urine samples from the Hospital were the same as those in the study group; these were, in order of greater to lesser prevalence, Escherichia coli with isolate percentages of 56% in the Hospital and 51% in the study group, Enterococcus faecalis with percentages of 18% and 11% respectively, Klebsiella pneumoniae with percentages of 7.9% and 11% respectively and Proteus mirabilis with percentages of 4.5% and 4% respectively. Of the 100 urine samples that comprised the study group, 63 were from women and 37 from men. The age of the patients was between 4 and 94 years, with an average age of 69 years; and 23 of them were hospitalized.
Results of identification obtained by MALDI-TOF from culture of positive urine samples from 2010 and from culture in the study group of 100 urine samples.
| Positive urines year 2010 (N=3272) (%) | Study group (N=100) (%) | |
|---|---|---|
| Enterobacteriaceae | 68 | 76 |
| NFGNRa | 7.9 | 4 |
| Staphylococcus spp. | 6.5 | 4 |
| Streptococcus spp. | 3.9 | 5 |
| Enterococcus spp. | 13.2 | 11 |
| Other microorganisms | 0.5 | 0 |
Urine samples with counts of ≥1×105CFU/ml produced higher identification scores (close to or above 2.000) in the majority of cases. On the other hand, the identification scores from urine with counts of <1×105CFU/ml were lower, such that identifications with scores <1.400 corresponded with microorganisms which are rarely etiological agents of urinary infection and, in numerous occasions, in one or more determinations of the four microorganisms carried out from the samples no peaks were obtained. Moreover, when scores were ≥1.400, despite the fact that the proposed microorganisms were included among the frequent UTIs source, and the identification proposed for each spot was different, the score was not a sufficient criterion for determining the identification of the bacteria, given that, in many cases, the suggestion with a higher score did not coincide with the identification obtained from culture. In this way, we observed that taking as valid the identification that was repeated at least in two of the four spots with counts of ≥1.400, we obtained 100% concordance in respect of the identifications carried out from culture.
Based on these data, we proposed the following validation criterion: a microorganism was correctly identified using MALDI-TOF when, after having taken four readings of the same sample, at least two readings offer the same identification proposal at species level for the first microorganism in the list, both with a score of ≥1.400.
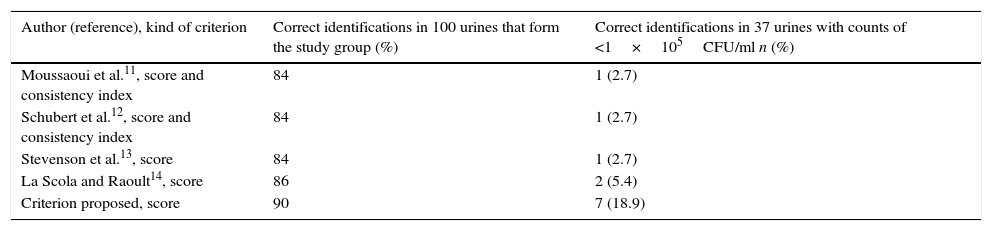
Identification results obtained from all urine samples processed in accordance with the validation criteria adopted are shown in Table 2. When the identification results obtained from the study group by applying the criterion proposed in this study were compared with those obtained by applying the validation criteria of different authors6–9, no statistically significant differences were observed (p>0.05). On the other hand, when the identification results from the 37 urine samples with counts of <1×105CFU/ml by applying the criterion proposed in this study were compared, we obtained statistically significant differences with respect to those obtained using the criteria of Moussaoui et al.6 (p<0.05), Schubert et al.7 (p<0.05) and Stevenson et al.8 (p<0.05); however, no statistically significant differences were observed (p>0.05) with respect to the criterion of La Scola and Raoult9.
Frequency of microorganisms identified by MALDI-TOF from 137 monobacterial urine samples related with the validation criterion adopted.
| Author (reference), kind of criterion | Correct identifications in 100 urines that form the study group (%) | Correct identifications in 37 urines with counts of <1×105CFU/ml n (%) |
|---|---|---|
| Moussaoui et al.11, score and consistency index | 84 | 1 (2.7) |
| Schubert et al.12, score and consistency index | 84 | 1 (2.7) |
| Stevenson et al.13, score | 84 | 1 (2.7) |
| La Scola and Raoult14, score | 86 | 2 (5.4) |
| Criterion proposed, score | 90 | 7 (18.9) |
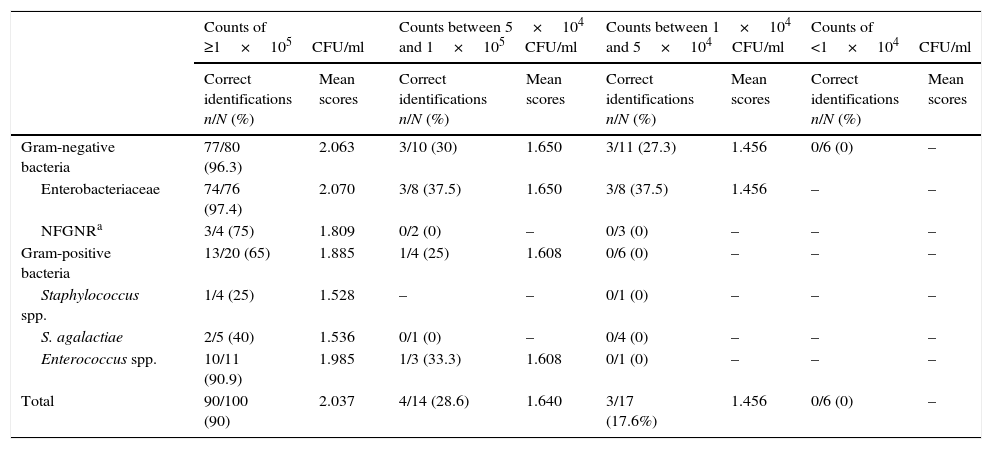
Bacterial identification results obtained by MALDI-TOF from 137 urine samples by applying the validation criterion proposed in this study are shown in Table 3. Of the 51 samples with counts of ≥1×105CFU/ml in which E. coli was identified from culture, there was only one failed identification from direct urine. The eleven isolates of K. pneumoniae, the four of P. mirabilis and Klebsiella oxytoca, the three of Citrobacter freundii, and the one of Enterobacter aerogenes and Serratia marcescens identified from culture were correctly identified from urine. Finally, of the two samples in which Enterobacter cloacae was isolated, correct identification was obtained in one of them. Of the four samples in which non-fermenting Gram-negative rods (NFGNR) were identified (two urine samples with Pseudomonas aeruginosa, one with Acinetobacter baumannii and one with Acinetobacter genomospecies 3), there was only one failed identification, which corresponded to a urine with P. aeruginosa. Staphylococci were isolated in four samples (two urine samples with Staphylococcus aureus, one with Staphylococcus epidermidis and one with Staphylococcus saprophyticus); in this set of four samples, only one correct direct identification was obtained, which corresponded to one isolate of S. aureus. E. faecalis was isolated from eleven samples, and from ten of them correct identification was obtained from urine directly. In the same way, two correct identifications were obtained from a set of five samples with Streptococcus agalactiae. In this way, a 90% sensitivity was obtained (confidence interval of 95%: 81.96–94.84).
Frequency of microorganisms identified by MALDI-TOF from direct urine samples related with bacterial concentration applying the criterion proposed in this study.
| Counts of ≥1×105CFU/ml | Counts between 5×104 and 1×105CFU/ml | Counts between 1×104 and 5×104CFU/ml | Counts of <1×104CFU/ml | |||||
|---|---|---|---|---|---|---|---|---|
| Correct identifications n/N (%) | Mean scores | Correct identifications n/N (%) | Mean scores | Correct identifications n/N (%) | Mean scores | Correct identifications n/N (%) | Mean scores | |
| Gram-negative bacteria | 77/80 (96.3) | 2.063 | 3/10 (30) | 1.650 | 3/11 (27.3) | 1.456 | 0/6 (0) | – |
| Enterobacteriaceae | 74/76 (97.4) | 2.070 | 3/8 (37.5) | 1.650 | 3/8 (37.5) | 1.456 | – | – |
| NFGNRa | 3/4 (75) | 1.809 | 0/2 (0) | – | 0/3 (0) | – | – | – |
| Gram-positive bacteria | 13/20 (65) | 1.885 | 1/4 (25) | 1.608 | 0/6 (0) | – | – | – |
| Staphylococcus spp. | 1/4 (25) | 1.528 | – | – | 0/1 (0) | – | – | – |
| S. agalactiae | 2/5 (40) | 1.536 | 0/1 (0) | – | 0/4 (0) | – | – | – |
| Enterococcus spp. | 10/11 (90.9) | 1.985 | 1/3 (33.3) | 1.608 | 0/1 (0) | – | – | – |
| Total | 90/100 (90) | 2.037 | 4/14 (28.6) | 1.640 | 3/17 (17.6%) | 1.456 | 0/6 (0) | – |
Of the 37 urine samples with counts of <1×105CFU/ml direct identification was achieved in seven of them. Of the 14 urine samples with counts between 5×104 and 1×105CFU/ml there were ten with Gram-negative bacterial isolates, and from these three correct identifications were obtained, which were two isolates of E. coli and one of K. pneumoniae. Three isolates of E. coli, one of P. mirabilis and one of Morganella morganii were not identified from the sample directly. On the other hand, of the four samples with Gram-positive bacterial isolates (two urines with an isolate of E. faecalis, one with Enterococcus faecium and one with S. agalactiae) only one identification of E. faecalis was obtained.
Of the 17 urine samples with counts between 1×104 and 5×104CFU/ml there were 11 with Gram-negative bacterial isolates which provided three acceptable identifications of E. coli from a direct sample; three isolates of E. coli, two of P. mirabilis and three of P. aeruginosa were not identified. In the six samples with Gram-positive bacterial isolates no correct identification was obtained. In this case, the strains identified from culture were one of S. aureus, four of E. faecalis and one of S. agalactiae.
Of the six urine samples processed with counts of <1×104CFU/ml, no valid identification was obtained. Five strains of E. coli and one of P. mirabilis were identified from culture.
As can be seen in Table 3, as the bacterial concentration was reduced in the samples, the percentages of identification and average scores obtained were also reduced. For each urinary bacterial concentration, the bacteria belonging to the family Enterobacteriaceae showed the greater percentage of correct identifications and higher average score, and in the same way, the percentage of correct identifications and the average score in Gram-negative bacteria were greater than that of Gram-positive bacteria.
In the identification of yeasts by MALDI-TOF from four urine samples, the species identified from culture was Candida albicans. In the two urine samples with counts of ≥1×105CFU/ml identification was obtained with an average score of 1.989, and in the two with counts between 1×104 and 5×104CFU/ml no valid identification was obtained.
DiscussionThis paper was focused on the direct microbial identification in urine samples regardless of their concentration because, apart from the classic criterion described by Kass13, it had demonstrated that counts of <1×105CFU/ml of bacteria and yeasts might be indicative of UTI in samples obtained through midstream clean catch14–18.
The criteria for validation of results obtained using MALDI-TOF can be classified into two groups: one takes into account the score and consistency index obtained in each reading and the other is based only on the score of the first microorganism in the list. Belonging to the first group two proposals have been reported. So, Moussaoui et al.6 established that an identification is valid if a score of ≥1.400 is obtained for the first microorganism in the list and the species in the first four identification proposals coincides; similarly, Schubert et al.7 advocated that an identification is valid if a score of ≥1.500 is obtained for the first microorganism and the species is the same in the first three identification proposals in the list. Based on the score obtained for the first microorganism in the list given by the software, two criteria can be found in the literature, i.e., the one proposed by Stevenson et al.8, and subsequently by Prod’hom et al.19, Juiz et al.20 and Ferreira et al.21, establishing that a score of ≥2.000 in an identification indicates a species level identification, a score between 1.700 and 1.900 indicates a genus level identification, and a score of <1.700 indicates non-identification. Finally, the standard proposed by La Scola and Raoult9 established that a microorganism is identified correctly using MALDI-TOF when, after having performed four identifications of a single sample, the bacterial species with the highest score (the first on the list) coincides in all of them and a score of ≥1.900 is obtained in at least two identifications, or when a score of ≥1.200 is obtained in the four identifications.
Urine samples with counts of ≥1×105CFU/ml of Gram-negative bacteria and Enterococcus spp. provided high identification scores (close to 2.000) and therefore high correct identification percentages were obtained, regardless of the validation criteria used. It is worth mentioning that the proposed criterion produced more direct identification percentages in the study group (Table 2). However, for the urine samples with counts of ≥1×105CFU/ml of Staphylococcus spp. or Streptococcus spp. and urine with counts of <1×105CFU/ml, low identification scores (close to 1.400) were obtained, and as a consequence, low identification percentages were obtained. For these urine samples with low scores, the identification percentage obtained depended on the criterion used in validating results and our criterion also produced higher percentages of correct direct identification (Table 2). The criteria of Moussaoui et al.6 and Schubert et al.7, which take into account the score and the consistency index, were not suitable for the validation of these urine samples that provided low identification scores. It was due to the lack of concordance at species level on the list of the ten microorganisms proposed by the software, despite the fact that the first option in the list could coincide with the identification obtained from culture. La Scola and Raoult9, who used lower scores than all those published to date, proposed substituting the consistency index obtained in a reading with the concordance at species level obtained in the first identification of the four readings carried out from the same sample; which is to say, this criterion is based on the score and the reproducibility of the equipment. A limitation of this criterion consists in that proposing valid identification from scores of ≥1.200 is not useful for validating of the results obtained from direct urine samples with low bacterial counts, since we have observed that scores of <1.400 suggested microorganisms which are rarely etiological agents in UTI. Using the criterion proposed in this study, the higher identification score obtained in one of the four readings carried out from one sample was not a determining factor, given that if an identification with a higher score than any other of those repeated was obtained, this should be discarded. So, with the proposed criterion, the weighting of the score was reduced in the validation of results.
When 5ml of urine with counts of ≥1×105CFU/ml of Staphylococcus spp. and Streptococcus spp. or with counts of <1×105CFU/ml was processed, the bacterial concentration was insufficient for carrying out direct identification by MALDI-TOF from urine. This suggested that in order to achieve the direct identification successfully it would be better to process greater sample volumes in order to process a greater amount of bacteria.
No previous studies on direct identifications of yeasts from urine samples have been found in the literature. Although the number of samples studied in which a yeast was isolated is very low, the results obtained coincided with those from bacteria positive urine with regard to the microbial concentration in the sample; which is to say, the two strains of C. albicans present in two urine samples with counts of ≥1×105CFU/ml were correctly identified by applying our criterion and the two strains isolated in two samples with counts of <1×105CFU/ml were not.
Results obtained in this study concur with those of Ferreira et al.21 and with those of Kohling et al.22, which also observed that counts of ≥1×105CFU/ml produced higher percentage of direct identification in urine samples, and that concentrations <1×105CFU/ml produced low percentages of direct identification. Our results also matched with those published by Christner et al.23, who pointed out that with greater bacterial concentration in the sample, greater is the quantity of microorganisms deposited on the metal sample plate and consequently a higher score was obtained. In this way, when the bacterial concentration in the urine sample is high (≥1×105CFU/ml) satisfactory identification percentages are obtained, above all in Gram-negative bacteria. In urine with counts between 5×104 and 1×105CFU/ml identification percentages were considerably reduced; in urine with counts between 1×104 and 5×104CFU/ml the percentages were reduced even further; and in urine with counts of <1×104CFU/ml, no direct identification was achieved. Moreover, for each urine bacterial concentration the average score for Gram-negative bacteria obtained is superior to that of Gram-positive bacteria (Table 3); these data coincides with that published by Schubert et al.7 who asserted that the MALDI-TOF system offers greater identification capacity for Gram-negative than Gram-positive bacteria.
For performing bacterial identification directly from urine samples, the clinician must request it in patients with suspected severe UTI. Direct bacterial identification cannot be applied in all urine samples because they are those most commonly processed in clinical microbiology laboratories24.
Direct identification is reported in less than one hour from the arrival of the urine to the laboratory and only includes the bacterial species present in urine. For confirmation of direct identification and clinical significance, it is necessary to perform bacterial identification and calculate bacterial counts from colonies obtained on isolation plates. Therefore, the clinician must assess clinical significance of direct bacterial identification and adopt appropriate measures regarding antimicrobial therapy.
A problem of direct identification by MALDI-TOF is that polymicrobial samples could provide wrong identifications since the spectra obtained include peaks of two or more microorganisms. This problem has been partially solved with the use of software (Bruker DaltoniK GmbH, Bremen, Germany) which, in some cases, is able to report two bacterial identifications if both microorganisms are in correct proportions.
To sum up, applying the differential centrifugation protocol together with the validation criterion proposed in this study, it was possible to achieve identification by MALDI-TOF of the microorganisms that most frequently cause UTI from direct monomicrobial urine samples with bacterial counts of ≥1×105CFU/ml; and, it was also possible to improve correct identifications in urine samples with bacterial counts of <1×105CFU/ml.
Conflict of interestThe authors declare no conflict of interest.