Shiga toxin-producing Escherichia coli (STEC) are a heterogeneous group of foodborne pathogens causing a broad spectrum of human disease, from uncomplicated diarrhea to hemolytic uremic syndrome (HUS). In this study, we report an HUS case associated with an O59:NM[H19] strain, harboring stx2a, iha, lpfAO26, lpfAO113 genes associated with STEC, and aatA, aap, pic, sigA, agg4A genes associated with enteroaggregative E. coli (EAEC), named Stx-EAEC. The strain showed low toxicity on Vero cells, and was resistant to streptomycin and trimethoprim/sulfonamides. The child carried the bacteria for more than 100 days. Since the large outbreak associated with Stx-EAEC O104:H4, many strains with similar profiles have been described. In Germany, an O59:NM[H19] strain, with comparable characteristics to the Argentine strain, was isolated from a bloody diarrhea case. In Argentina, this is the first report of an HUS case associated with a Stx-EAEC infection, and represents a new challenge for the surveillance system.
Escherichia coli productor de la toxina Shiga (STEC) es un grupo heterogéneo de patógenos transmitidos por alimentos que causan un amplio espectro de enfermedades humanas, desde diarrea no complicada hasta síndrome urémico hemolítico (SUH). Nosotros informamos de un caso de SUH por O59:NM[H19], que portaba los genes stx2a, iha, lpfAO26, lpfAO113 asociados con STEC, y los genes aatA, aap, pic, sigA, agg4A de E. coli enteroagregativo (EAEC), llamado EAEC-Stx. La cepa mostró baja citotoxicidad en las células Vero, y fue resistente a estreptomicina y trimetoprima/sulfonamidas. El niño excretó la bacteria durante más de 100 días. Desde el brote asociado con EAEC-Stx O104:H4, se describieron muchas cepas con perfiles similares. En Alemania se aisló una cepa O59:NM[H19] de una diarrea sanguinolenta, con características comparables a la cepa argentina. Este es el primer informe de un caso de SUH asociado a una infección por EAEC-Stx, y representa un nuevo desafío para el sistema de vigilancia.
Shiga toxin (Stx)-producing Escherichia coli (STEC) are a heterogeneous group of foodborne pathogens causing diarrhea, hemorrhagic colitis and hemolytic uremic syndrome (HUS). Argentina has the highest incidence of HUS globally, with approximately 400 new cases each year. During the period 2005–2015, the incidence in children under 5 years of age was 8.5 cases per 100000 annually. Evidence of STEC infection is found in around 70% of HUS cases and O157:H7 is the predominant serotype isolated8. In 2011, a STEC O104:H4 strain caused a large outbreak in Germany with more than 4000 infections, including 900 HUS cases and 50 deaths2. The outbreak strain, besides the production of Stx, possessed the adhesion determinants of Enteroaggregative E. coli (EAEC). This new pathotype was named as Enteroaggregative Hemorrhagic E. coli (EAHEC)1,4, hybrid of EAEC/STEC9 and Stx-EAEC3 by different authors, in order to better define the virulence profile.
In this report, we describe the isolation and characterization of a Stx-EAEC strain that was associated with an HUS case in Argentina and compare the strain to similar strains isolated in Germany.
Clinical and laboratory findingsOn January 15, 2015, a boy who was 7 years and 8 months old was seen by his pediatrician for fever (38°C), and vomiting. The child presented severe headache, irritability, photophobia, abdominal pain, but no upper tract catarrh, and was sent home with dietary and general instructions. Only a nonsteroidal anti-inflammatory drug was given as treatment. As the general condition worsened, the next day he was seen by his pediatrician again, who sent him to the emergency room of Hospital Zonal de Puerto Madryn “Dr. Andrés Ísola”. Upon admission, he exhibited generalized pallor, jaundice of skin and mucous membranes, an episode of loose stool, dehydration, arterial hypertension (blood pressure 150/90mmHg [greater than 95th percentile of systolic blood pressure for age]) and sleepiness, and, given his condition, he was hospitalized in the intensive care unit (ICU). In the ICU, the child had oliguria and dark urine, with an initial diagnostic impression of acute renal failure, anemia and thrombocytopenia. A presumptive diagnosis of atypical HUS (D-) was established. Initial laboratory findings included: hematocrit, 28%; hemoglobin level, 9.2g/dl; leukocyte count, 10700/mm3; platelet count, 33000/mm3; serum glucose, 106mg/dl; blood urea nitrogen (BUN), 206mg/dl; serum creatinine, 1.9mg/dl; albumin, 4.1g/dl; uric acid, 7.5mg/dl; calcium, 9.4mg/dl; phosphorus, 6.5mg/dl; magnesium, 2.8mg/dl; sodium, 134mEq/l; potassium, 4.4mEq/l; pH, 7.36; bicarbonate, 16mmol/l; base excess, −8mmol/l; AST, 133U/l; ALT, 16U/l; total bilirubin, 4mg/dl; direct bilirubin, 1mg/dl; indirect bilirubin, 3mg/dl; prothrombin time, 12.7s; lactic acid, 1.1mmol/l; lactate dehydrogenase (LDH), 5130UI/l. A stool culture was requested. During hospitalization, the patient received hydration, omeprazole, nifedipine, allopurinol, calcium carbonate, bicarbonate, and three infusions of packed red blood cells, but did not require dialysis. Antimicrobial therapy was not provided. The blood and urine cultures were negative. On the 8th day, he had a seizure episode with paresis of the left hemisphere that improved with lorazepam treatment. He continued the treatment with clobazan, and remained with left hemiparesis and ataxia that improved in the following days. Cranial magnetic resonance imaging (MRI) and X-ray computed tomography (CT) scans showed normal parameters. The patient was discharged after 11 days of hospitalization. Since the hospital discharge and until now, he has been periodically controlled by a pediatric nephrologist, confirming on February 2018 absence of proteinuria and microalbuminuria and normal renal function.
Epidemiological investigationAs part of the epidemiological study conducted at the hospital to identify the risk factors associated with the disease, the parents were interviewed using a standardized questionnaire. Information was collected about potential exposure in the 7 days before the onset of symptoms, and demographic issues. On January 11, the child and his family (parents and a 42-months-old sister) had eaten well-done roast meat and sausages purchased at a local market. No other family members reported gastrointestinal symptoms. The parents referred good practices of food handling and hygiene in the home. Informed consent was obtained from the adult primary caregiver, who was interviewed in person.
Microbiological investigationA stool sample was collected on January 16 and routinely cultured for E. coli, Salmonella, and Shigella species. The stool sample and a culture on MacConkey (MAC) agar (Difco Laboratories, Detroit, MI) were sent to the Servicio Fisiopatogenia, as National Reference Laboratory (NRL), for STEC and free fecal Stx (FFStx) detection, as previously described11. The material was sent with the respective hospital charts (records), from which the clinical, laboratory and epidemiological data were reviewed. The bacterial confluent growth zone on MAC was positive for stx2 gene by a multiplex polymerase chain reaction (mPCR)11. The isolate (EC-90 #1) identified as E. coli by standard biochemical methods, was defined as non-O157/stx2 by the mPCR. The strain was negative for eae (intimin encoded protein) and ehxA (enterohemolysin) genes by PCR11. FFStx2 was detected by specific cytotoxicity assays on Vero cells.
From a second stool sample, collected on March 17, 61 days after the onset of symptoms, E. colistx2-positive (EC-90 #2) and FFStx2 were detected. Due to the prolonged STEC shedding, virulence factors of other diarrheagenic E. coli pathotypes were analyzed by PCR13. Both isolates (EC-90 #1 and EC-90 #2), corresponding to the first and second samples, were positive for the regulator AggR, which drives the expression of AraC, and also the expression of the different aggregative adherence fimbriae (AAF) responsible of the characteristic stacked brick-like adherence pattern of EAEC and other proteins3. A third E. coli strain (EC-90 #3) with the same virulence profile was recovered from feces 115 days after the onset of symptoms and the stool sample was also FFStx2-positive.
The full characterization done by PCR as previously described7,9,14,15, showed that the three strains harbored stx2a (Shiga toxin variant 2a), iha (gene for IrgA adhesion homolog), lpfAO26 (gene for structural subunit of long polar fimbriae (LPF) of STEC O26), lpfAO113 (gene for structural subunit of LPF of STEC O113) genes frequently associated with STEC, and aatA (dispersin transporter protein), aap (dispersin, antiaggregation protein), pic (protein involved in intestinal colonization), sigA (gene for the SPATE protein) genes typically associated with EAEC. The presence of type IV AAF was established by the amplification of the agg4A fimbrial subunit gene. The EC-90 #1, EC-90 #2 and EC-90 #3 strains, were phenotypically serotyped as O59:NM and the fliCH19 gene was determined by PCR, using primers described previously5. The strains showed a cytotoxicity of 600 CD50/90μl on Vero cells, and were resistant to streptomycin and trimethoprim/sulfonamides. Stool samples collected at 145 and 162 days were negative for Stx-EAEC and FFStx.
DiscussionUntil recent years, the ability to produce Stx and to cause the attaching-and-effacing (A/E) lesion was the paradigm for severe HUS-associated STEC. The Stx-EAEC O59:NM[H19] strain, described in the present study, had the ability to produce HUS and to cause a long-term shedding in the patient. Although the strain showed low cytotoxicity on Vero cells, its pathogenicity and prolonged excretion could be attributed to the fact that it has characteristics of EAEC; therefore it is a better colonizer of the intestine due to its aggregative phenotype. This property could facilitate the systemic absorption of Stx.
Since the large German outbreak associated with a Stx-EAEC O104:H4 strain, attention has been focusing on this new E. coli pathotype. Prager et al. screened 2435 strains of the STEC collection from the German National Reference Centre for Salmonella and other Bacterial Enteric Pathogens (NRC), corresponding to the period 2008–2012, for the presence of stx1, stx2, eae and ehxA genes10. Among 268 eae- and ehxA-negative strains, two strains exhibited both STEC and EAEC marker genes and were stx2- and aatA-positive. One strain, isolated from a case of bloody diarrhea in 2010 and serotyped as O59:NM[H19], harbored stx2a, belonged to the sequence type (ST) 1136 by multiple locus sequence typing (MLST), and exhibited genes for type IV aggregative AAF fimbriae, with resistance toward sulfonamides, streptomycin, and trimethoprim/sulfonamide and low cytotoxicity on Vero cells.
The German O59:NM[H19] strain harbored the same genes as the Argentine O59:NM[H19] strain plus the following genes: irp2 (component of iron uptake system on high pathogenicity island) gene of STEC, and set1a (Shigella enterotoxin subunit A), set1b (Shigella enterotoxin subunit B), iucA (siderophore aerobactin) genes of EAEC. The second German strain, isolated from a patient with diarrhea in 2012, harbored stx2b, was typed as Orough:H2 and belonged to MLST ST26.
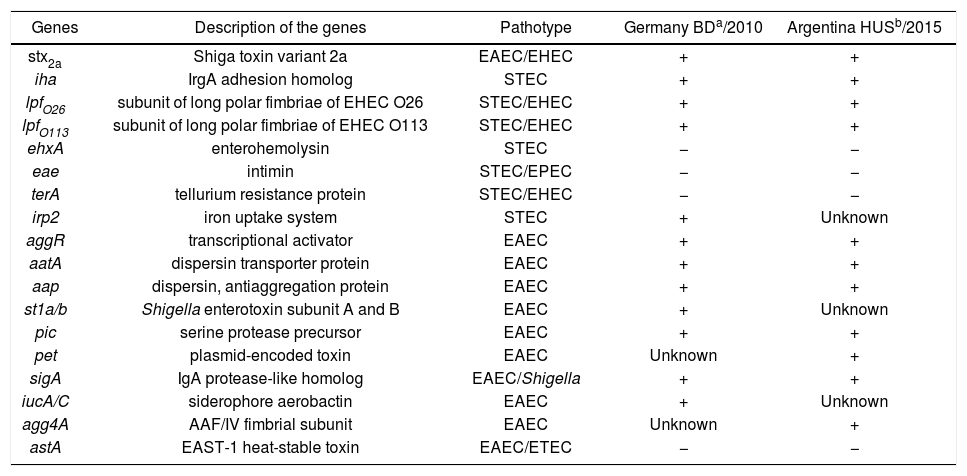
A comparison between the properties of both Stx-EAEC O59:NM[H19] strains is shown in Table 1.
Properties of Stx-EAEC O59:NM[H19] strains isolated in Germany and Argentina
| Genes | Description of the genes | Pathotype | Germany BDa/2010 | Argentina HUSb/2015 |
|---|---|---|---|---|
| stx2a | Shiga toxin variant 2a | EAEC/EHEC | + | + |
| iha | IrgA adhesion homolog | STEC | + | + |
| lpfO26 | subunit of long polar fimbriae of EHEC O26 | STEC/EHEC | + | + |
| lpfO113 | subunit of long polar fimbriae of EHEC O113 | STEC/EHEC | + | + |
| ehxA | enterohemolysin | STEC | − | − |
| eae | intimin | STEC/EPEC | − | − |
| terA | tellurium resistance protein | STEC/EHEC | − | − |
| irp2 | iron uptake system | STEC | + | Unknown |
| aggR | transcriptional activator | EAEC | + | + |
| aatA | dispersin transporter protein | EAEC | + | + |
| aap | dispersin, antiaggregation protein | EAEC | + | + |
| st1a/b | Shigella enterotoxin subunit A and B | EAEC | + | Unknown |
| pic | serine protease precursor | EAEC | + | + |
| pet | plasmid-encoded toxin | EAEC | Unknown | + |
| sigA | IgA protease-like homolog | EAEC/Shigella | + | + |
| iucA/C | siderophore aerobactin | EAEC | + | Unknown |
| agg4A | AAF/IV fimbrial subunit | EAEC | Unknown | + |
| astA | EAST-1 heat-stable toxin | EAEC/ETEC | − | − |
a,b Both strains, isolated from bloody diarrhea (BD) and hemolytic uremic syndrome (HUS) cases, showed resistance to streptomycin and trimethoprim/sulfamethoxazole and low cytotoxicity on Vero cells.
By pulsed-field gel electrophoresis using the restriction enzyme XbaI (XbaI-PFGE), the three Argentine strains showed the same macrorestriction pattern, named AREEF901.0001, with 100% similarity11. A high clonal relationship (>85%) with the Stx-EAEC O59:NM[H19] German strain was established. The dendrogram of the German and Argentine strains is shown in Figure 1.
The impact of the German outbreak was so important to become the responsible E. coli strain as a prototype of the new Stx-EAEC pathotype. The emergence of Stx-EAEC O104:H4 suggests that either certain EAEC serotypes might be more susceptible to acquire STEC determinants or that there are certain Stx-EAEC ancestors which successfully adapted to survive specific selection conditions, which represents a new challenge for the entire scientific community. There is evidence that shows that the Stx-EAEC O104:H4 could have evolved by the uptake of a stx2-phage originated from the bovine reservoir by an EAEC O104:H4 strain1.
Other Stx-EAEC strains of different serotypes were previously described in association with human disease. During an HUS outbreak in France in 1996, an O111:H2 strain was characterized as stx2- and AAF-positive, and eaeA- and ehxA-negative, being the first Stx-EAEC hybrid described. In 1999, an O86:NM[H2] strain with stx2 and AAF marker genes but no eae was isolated from a child with HUS and bloody diarrhea in Japan. Later, in 2011, an O111:H21 strain associated with a household outbreak in Northern Ireland was detected. The strain harbored the stx2c gene and the type V AAF fimbriae, but was eae-negative with a low level of resistance to ampicillin6. In 2013 in a school in northern Italy, an outbreak caused by a Stx-EAEC- O127:H4 strain was reported. Five Stx-EAEC strains were isolated from 76 fecal samples from children and school staff. The strains were characterized as O127:H4 and ST678 and possessed the stx2a gene and other EAEC virulence genes such as aggR, aap, aat, aaiC, sigA, pic and astA. The stx2-phage was inserted in wrbA site, as well as it was described for O104:H4 outbreak strain12.
Tozzoli et al. have demonstrated that, at laboratory level, all stx2-phages used in their study were able to infect a range of bacterial hosts, including both diarrheagenic (DEC) and extraintestinal pathogenic E. coli (ExPEC), suggesting that STEC might represent multiple pathogroups whose virulence has been increased by the event of a stx-phage acquisition16. Regarding the Stx-EAEC, it was demonstrated by genomics sequencing, that EAEC strain was infected by stx2-phage and yield this new pathotype3. On the other hand Nyholm et al. also described hybrid Shigatoxigenic and Enterotoxigenic Escherichia coli (STEC/ETEC) strains, and 1% of the strains belonging to a STEC collection harbored genes typically present in ETEC and some of them being isolated from patients with HUS9.
In Argentina, this is the first report of an HUS case associated with a Stx-EAEC infection. A distinct characteristic was the prolonged shedding during 4 months. Although O59:NM[H19] is an unusual serotype in the country, these results open new questions and pose a challenge for the Surveillance System.
Conflict of interestThe authors declare that they have no conflicts of interest.
We thank Dr. Rita Prager, from the Division of Enteropathogenic Bacteria and Legionella, National Reference Center for Salmonella and other Bacterial Enteric Pathogens, Robert Koch-Institute, Wernigerode, Germany, for kindly sending us the TIFF of the German strain.





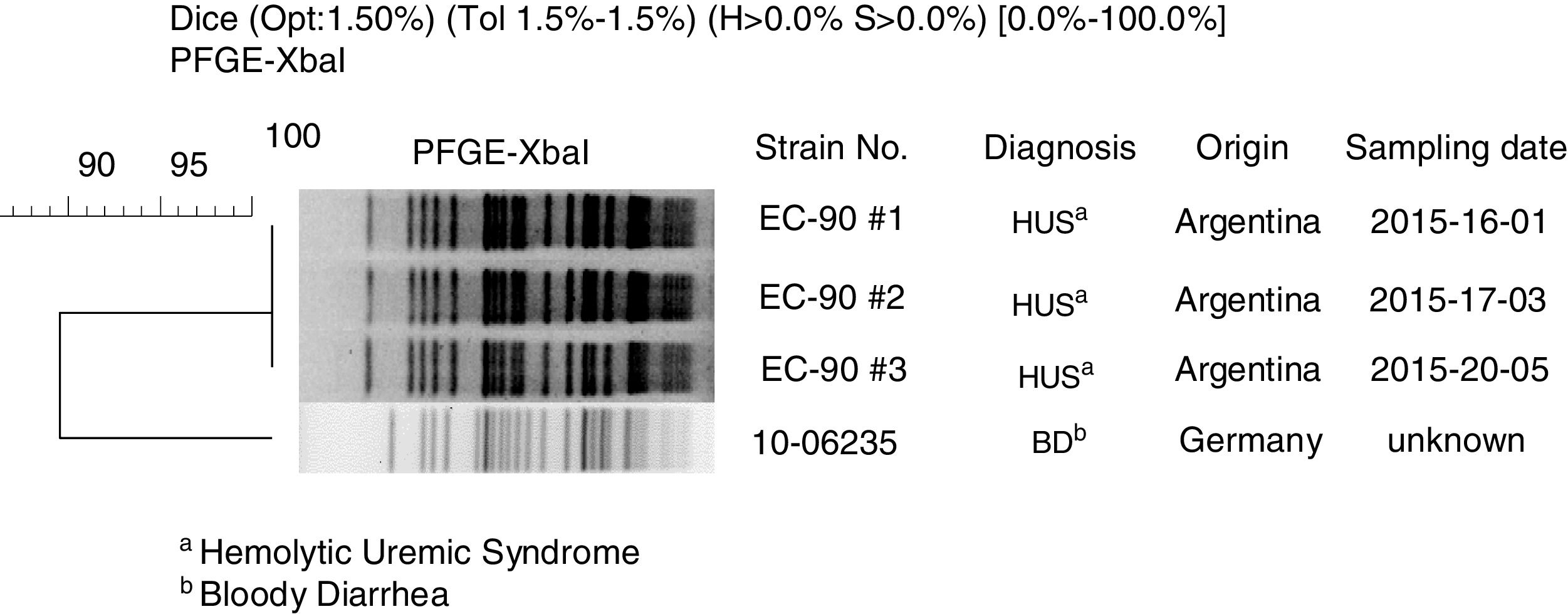
![Clonal relation between Stx-EAEC O59:NM[H19] strains isolated in Germany and Argentina by XbaI-PFGE. Clonal relation between Stx-EAEC O59:NM[H19] strains isolated in Germany and Argentina by XbaI-PFGE.](https://static.elsevier.es/multimedia/03257541/0000005200000001/v1_202003120635/S0325754119300525/v1_202003120635/en/main.assets/thumbnail/gr1.jpeg?xkr=ue/ImdikoIMrsJoerZ+w96p5LBcBpyJTqfwgorxm+Ow=)
