The goal of this study was to isolate, select and characterize bacteria with cellulolytic activity from two different coffee residue composting piles, one of which had an internal temperature of 57°C and pH 5.5 and the other, a temperature of 61°C, and pH 9.3. Culture media were manipulated with carboxymethylcellulose and crystalline cellulose as sole carbon sources. The enzyme activity was assessed by hydrolysis halo formation, reducing sugar production and zymograms. Three out of twenty isolated strains showed higher enzymatic activity and were identified as Bacillus subtilis according to their morphological, physiological, biochemical characteristics and based on the sequence analysis of 16S rDNA regions. The enzymatic extracts of the three selected strains showed exocellulase and endocellulase maximum activity of 0.254 and 0.519 U/ml, respectively; the activity of these enzymes was maintained even in acid pH (4.8) and basic (9.3) and at temperatures of up to 60°C. The enzymatic activities observed in this study are within the highest reported for cellulose produced by bacteria of the genus Bacillus. Endocellulase activity was shown in the zymograms from 24h until 144h of incubation. Furthermore, the pH effect on the endocellulase activity is reported for the first time by zymograms. The findings in this study entail the possibility to use these enzymes in the procurement of fermentable substrates for the production of energy from the large amount of residues generated by the coffee agroindustry.
El objetivo de este estudio fue aislar, seleccionar y caracterizar bacterias con actividad celulolítica a partir de 2 diferentes pilas de compostaje de residuos de café, una con temperatura interna de 57°C y pH 5,5; la otra con temperatura interna de 61°C y pH 9,3. Se utilizaron medios de cultivo con carboximetilcelulosa y celulosa cristalina como únicas fuentes de carbono. La actividad enzimática fue evaluada por formación de halos de hidrólisis, producción de azúcares reductores y zimogramas. De 20 cepas aisladas, 3 presentaron mayor actividad enzimática y fueron identificadas como Bacillus subtilis sobre la base de sus características morfológicas, fisiológicas y bioquímicas y del análisis de las secuencias de la región 16S del ADNr. Los extractos enzimáticos de las 3 cepas seleccionadas presentaron actividad de exocelulasa y de endocelulasa, con máximos de 0,254 y 0,519U/ml, respectivamente; la actividad de estas enzimas se mantuvo incluso a pH ácido (4,8) o básico (9,3) y a temperaturas de hasta 60°C. Las actividades enzimáticas halladas en este estudio se ubican dentro de las más altas reportadas para celulasas producidas por bacterias del género Bacillus. En los zimogramas se demostró actividad de endocelulasa desde las 24h hasta las 144h de incubación. Asimismo, se reporta por primera vez el efecto del pH sobre la actividad de endocelulasa observado por zimogramas. Los resultados de este estudio abren la posibilidad de hacer uso de estas enzimas en la obtención de sustratos fermentables para la producción de energía a partir de los residuos generados en grandes cantidades por la agroindustria del café.
The production of commercial microbial cellulases has a worldwide market. Cellulases are widely used in research and in the food, paper, textile, chemical and energy industries9,47. Most cellulases are derived from fungi because they have shown high activity and produced cellulases in large amounts. In recent years, the need for more robust cellulases in complex substrates, such as agricultural residues with high cellulose content, has increased due to their uses as potentially fermentable substrates in the production of biofuels20,30. One option for this would be bacterial cellulases, which form synergistic multi-enzymatic complexes with increased activity43. Moreover, bacterial cellulases are active at extreme pH, salinity and temperature conditions, such as those encountered in composting processes41, including high temperature environments where the presence of thermostable enzymes increase bioconversion rates1. The bacterial species that dominate the thermophilic phase of compost processes belong to the genus Bacillus. They play an important role in the degradation of complex substrates, such as cellulose10. There are several reports of bacterial cellulases with activities at extreme pH (acidic or basic), high salinity and temperatures3,6,17,18,24. These cellulases have been isolated from several environments, such as termite guts5, soil contaminated with paper industry residues37, mangrove soil7, degraded lignocellulosic biomass49 and compost systems of kitchen and garden residues, municipal solid wastes, coffee residues and other agricultural residues16,29,34,39,42. These cellulosic residues are complex, and each of them has characteristics conferring certain specificity. Residues generated by the coffee industry also contain caffeine, gallic acid, caffeic acid, chlorogenic acid, and phenolic compounds8 and have high acidity. In the Soconusco region, Chiapas, Mexico, large amounts of residues are generated in wet coffee processing (approximately 7200tons), equivalent to 80% of the annual coffee production of 9000tons46. These residues can be used to obtain fermentable substrates for the sustainable production of ethanol in coffee-producing zones. Thermostable and acidophilic enzymes are required to take advantage of the cellulose of complex substrates, such as those from this agricultural residue. Therefore, the aim of this study was to isolate, select and characterize bacterial strains with cellulolytic capacity from the composting process with coffee residues.
Materials and methodsIsolation of cellulolytic bacteria from composting with coffee residuesSamples were obtained from the middle area of two composting piles, each one containing coffee residues (pulp and husk) and cow manure at a ratio of 3:1 (v/v). The piles showed different characteristics: pile 1: nine days of composting, with an internal temperature of 57°C and pH 5.5; and pile 2: 36 days of composting with an internal temperature of 61°C and pH 9.3.
For the isolation of cellulose degrading bacteria, about 1g of sample was added to 9ml NaCl 0.9% and serially diluted and pour plate and spread plate techniques were done using mineral medium (g/l): 1.4 (NH4)2SO4, 2.0 KH2PO4, 0.3 urea, 0.3 CaCl2·2H2O, 0.3 MgSO4·4H2O, 0.005 FeSO4·7H2O, 0.0016 MnSO4·H2O, 0.0014 ZnSO4·7H2O, and 0.02 CoCl2·6H2O with different cellulosic materials of carbon source: (1) 10g/l carboxymethylcellulose (CMC), (2) 10g/l crystalline cellulose (CC), (3) 10g/l coffee waste (PC), (4) 9g/l CMC and 1g/l glucose (CMCG), (5) 9g/l CC and 1g/l glucose (CCG), (6) 9g/l PC and 1g/l glucose (PCG), (7) 10g/l CMC and 0.25g/l yeast extract (CMCYE), (8) 10g/l CC and 0.25g/l yeast extract (CCYE), and (9) 10g/l PC and 0.25g/l yeast extract (PCYE). All culture media were supplemented with 0.1% Congo red and 2% agar. The colonies that grew on the different culture media were isolated by streaking them on CMC media at pH 5.9, 7.0, and 9.3 and CC at pH 5.6 and 7.0. For the culture, and the isolation, the incubations were performed at 37°C for 5 days.
Selection of cellulolytic bacterial strainsThe bacterial strains that managed to grow in the CMC and CC media were re-grown in the CMCYE, pH 5.9 and CCYE, pH 5.6 liquid media, respectively, and were incubated at 37°C for 120h. At different time intervals, 200μl of the culture were sampled and evaluated to determine reducing sugar concentrations using dinitrosalicylic acid (DNS)32 to confirm cellulolytic capacity. A standard glucose curve was used to calculate the sugar concentrations. The strains were also inoculated in mineral media (g/l): 1.0 KH2PO4, 0.5 MgSO4·7H2O, 0.01 FeSO4·7H2O, 0.01 MnSO4·H2O, 0.3 NH4NO3 and 1% CMC at pH 5.6, 7.0, and 9.3. They were incubated at 37°C for 24h. The plates were stained with 0.5% Congo red for 15min, washed with 1M NaCl and left to rest for 10min4. Hydrolysis halo diameters were manually measured, and the strains with larger halos were selected.
Identification of bacterial strainsStrains with the highest cellulolytic activity were identified using biochemical and morphological analyses according to the Bergey's Manual of Systematic Bacteriology45 by scanning electron microscopy with a TOPCON SM-510 microscope and by analyzing 16S rDNA regions.
For the amplification of the ribosomal regions, the extraction of genomic DNA was performed for the bacterial strains using the phenol-chloroform technique44. For PCR, universal primers fD1 (5′-AGAGTTTGATCCTGGCTCAG-3′) and rD1 (5′-AAGGAGGTGATCCAGCC-3′)51 and Taq polymerase EPO402 (Fermentas) were used. The reaction was performed in a T100 thermocycler (BIORAD) under the following conditions: 95°C for 3.5min; followed by 30 cycles at 95°C for 30s, 53°C for 30s, and 72°C for 1.5min, with a final extension of 72°C for 10min. The products obtained were purified with Spin Quantum PCR Kleen columns (BIORAD) and sequenced in a capillary sequencer (Macrogen, Inc., Korea). Sequence analyses were performed with MEGA 6.0648 and with the NCBI Basic Local Alignment Search Tool (BLAST) algorithm. A redundancy analysis of previously aligned sequences with SINA36 was performed using DAMBE 5.252. Phylogenetic analyses were performed using the maximum likelihood statistical method with the Jukes–Cantor model19 using MEGA721. The phylogenetic tree was built based on the sequences of microorganisms reported in the LPSN bacterio.net26 reference database. Confidence was evaluated using 1000 bootstrap replicates.
Cellulase productionCultures were performed under the same selection conditions, strains were inoculated in triplicate in 250-ml Erlenmeyer flasks with 75ml of CMCYE, pH 5.9, and CCYE, pH 5.65, and incubated at 37°C at 200rpm. Every 24h, 5ml of the culture was taken and collected in 15ml conical tubes. They were then centrifuged at 10000rpm for 3min at 4°C to separate the biomass. Protease inhibitors (200μl cocktail CompleteTM ultra tablets, EDTA free and 50μl of 100mM PMSF) were subsequently added to the supernatant (protein extract). This protein extract was stored at −20°C until further analysis.
SDS-PAGE, Native-PAGE and zymogramsFor the analysis of proteins secreted by the bacterial strains, polyacrylamide gel electrophoresis under denaturing (SDS-PAGE) and native (Native-PAGE) conditions were performed with zymograms to determine cellulase activity22. For the SDS-PAGE, the protein extracts were concentrated by exposure to 99°C for 12min and then to −20°C for 16h. The precipitated proteins were resuspended in a 0.1% SDS/5.6M urea/50mMTris solution. For Native-PAGE, the electrophoresis was performed with crude protein extracts.
Loading volumes were established according to the amount of proteins, which were quantified by the Bradford technique12 using bovine serum albumin (BSA) as a standard. To assess the total protein profile, gels were stained with Coomassie blue R-250 for 16h.
For the zymograms, both SDS-PAGE and Native-PAGE gels with 0.1% CMC were incubated for 16h with different buffer solutions (0.1M citrate buffer at pH 4.8, 0.2M acetate buffer at pH 5.8, 1× PBS at pH 7.4 and 0.025M borate buffer at pH 9.3) at 25°C with mild agitation. The gels were stained with 0.5% Congo red and washed with 1M NaCl until bands appeared to reveal the cellulase activity38.
Enzymatic activity with CMC and CC at different pH and temperature valuesCellulase activity was defined as the amount of enzyme required to generate 1μmol of reducing sugars in one minute and was evaluated at 37°C and 60°C with 1% CMC and 1% CC in different buffers (0.1M citrate buffer at pH 4.8, 0.2M acetate buffer at pH 5.8, 1× PBS at pH 7.4 and 0.025M borate at pH 9.3) after 30min incubations. The concentrations of reducing sugars were quantified using the DNS method.32
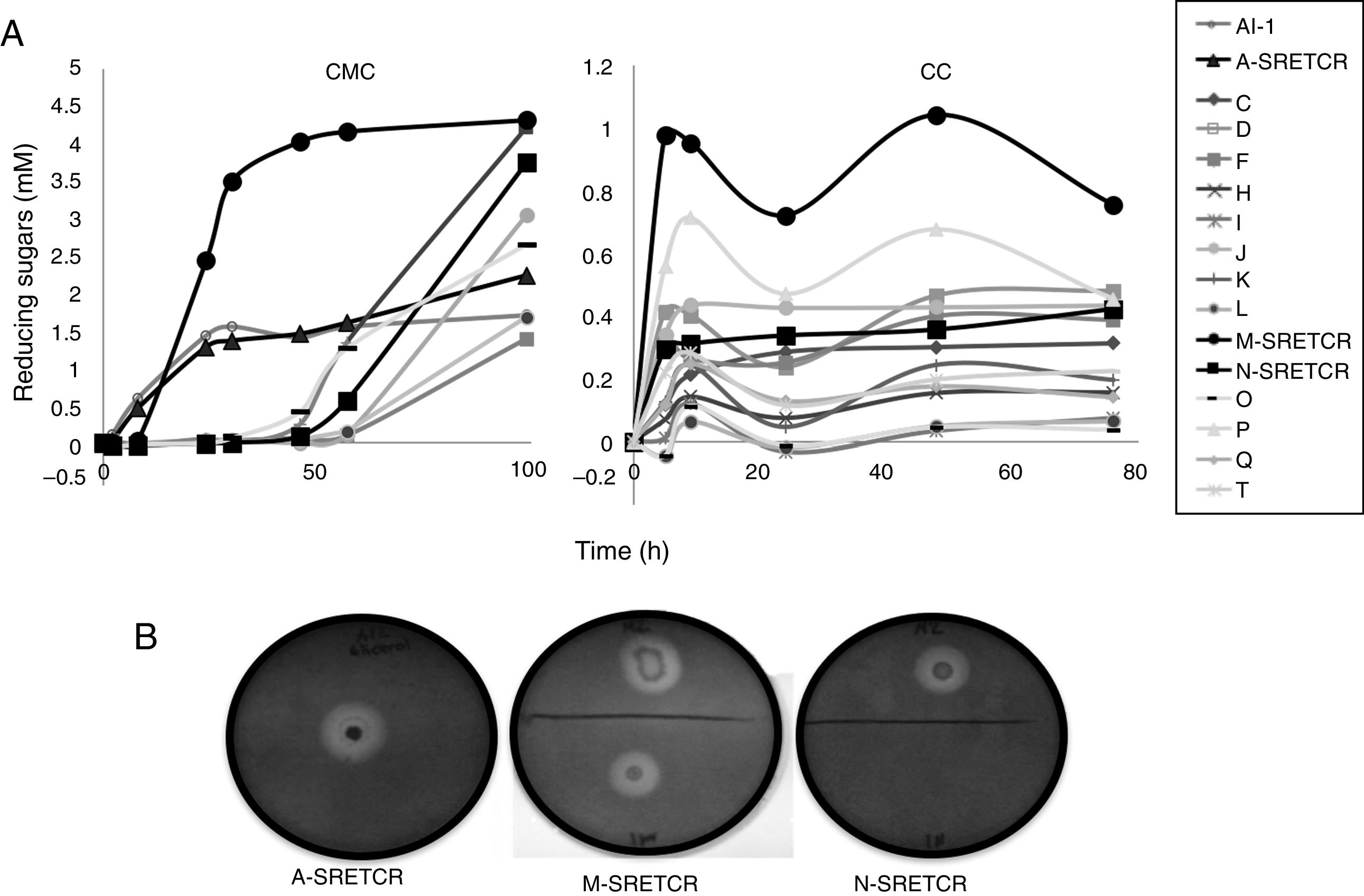
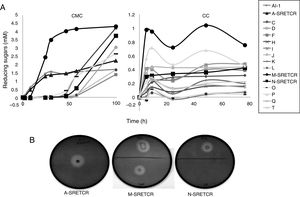
Results and discussionIsolation and selection of cellulolytic bacteria from compostingTwenty bacterial colonies were isolated from the samples obtained from the coffee residue composting piles; two colonies grew in the media with CMC, seven in the media with CC and seven in the media with both substrates. Although all strains released reducing sugars, the M-SRETCR strain released the highest concentration in both media: 2.4mM after 24h in CMC and 0.98mM after 5h in CC (Fig. 1A). The A-SRETCR strain produced sugars on the CMC media after 2h; the highest concentration (2.2mM) was observed after 99h; during the same 99h period, the M-SRETCR strain released 4.3mM. The A-SRETCR, M-SRETCR and N-SRETCR strains were the only bacteria that produced hydrolysis halos at pH 7.0 with diameters of 20, 18 and 15mm, respectively (Fig. 1B). These sizes were greater than those reported by Acharya et al.2 for B. subtilis isolated from compost piles. However, the halos reported by Acharya were produced after 8 days of incubation, whereas those obtained in this study were observed after 24h. The A-SRETCR and M-SRETCR strains also showed halos at pH 5.0 and 9.0, with diameters of 15 and 10mm, respectively.
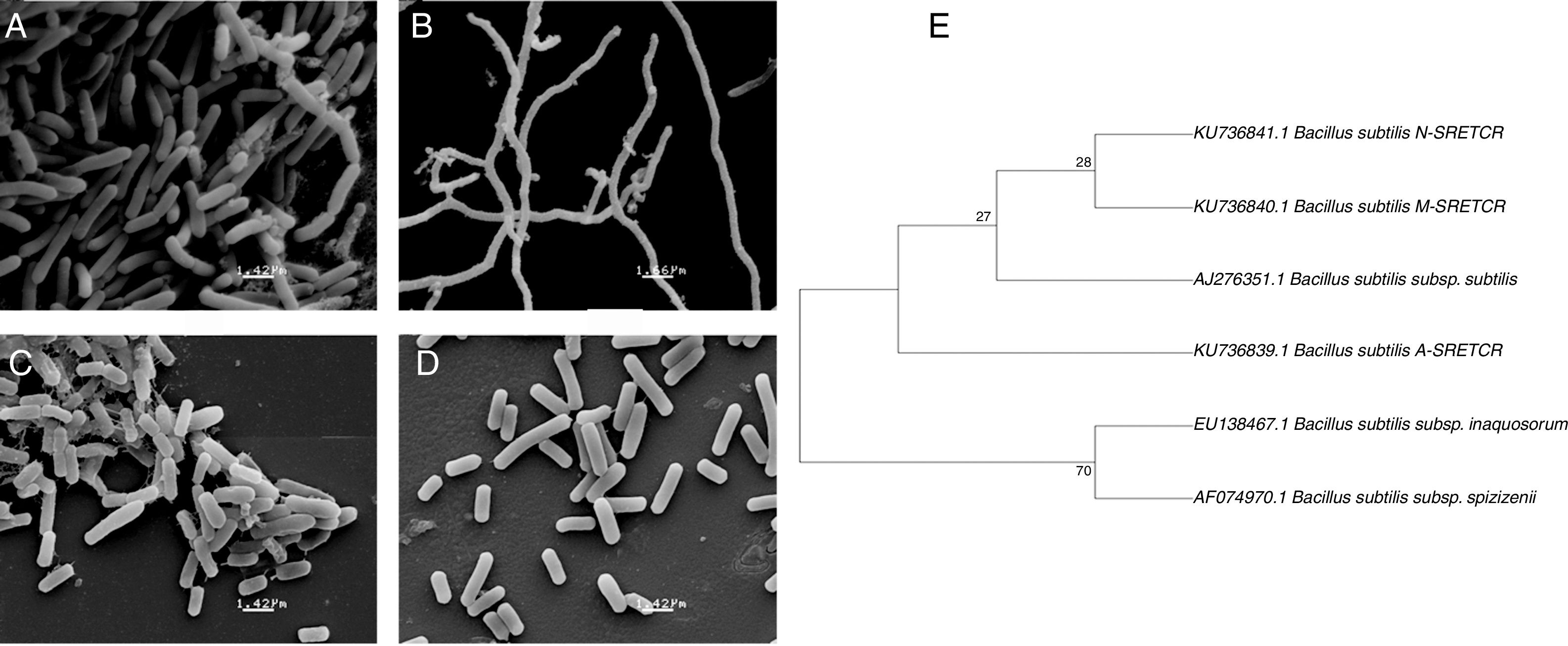
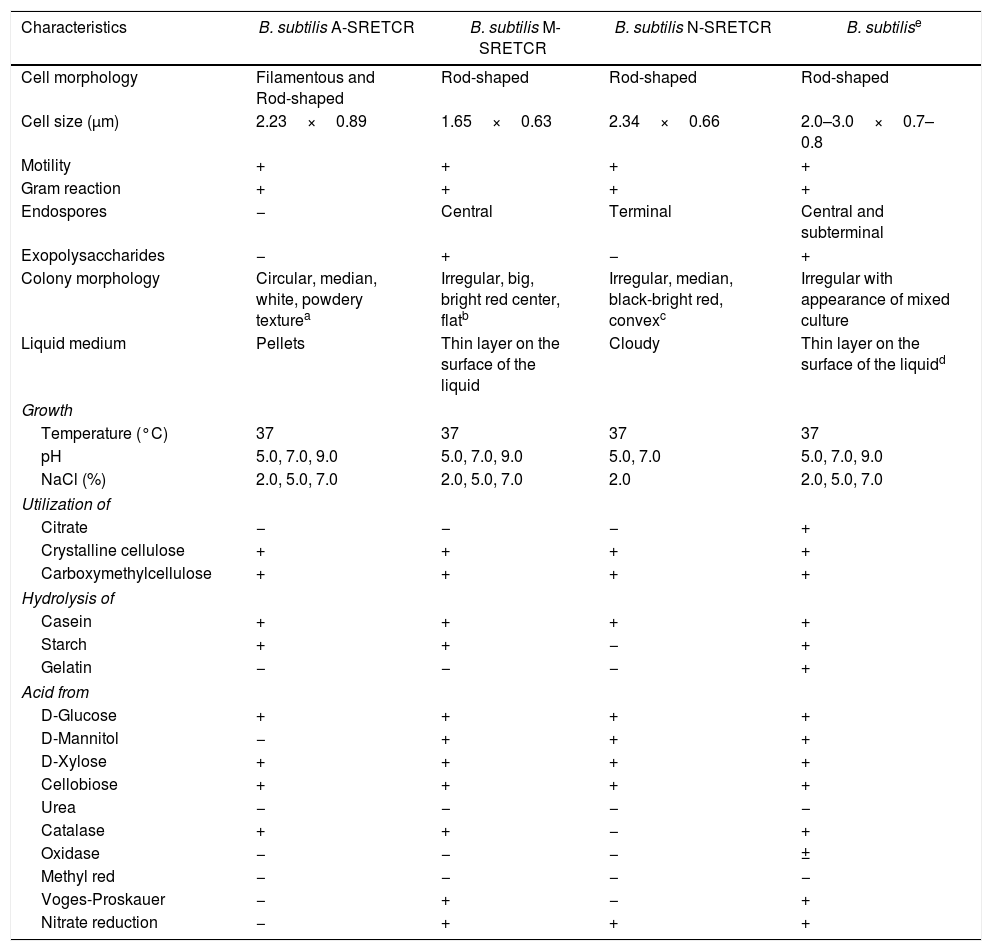
Identification of cellulolytic bacterial strainsThe A-SRETCR, M-SRETCR and N-SRETCR strains were selected due to their higher cellulolytic activity and identified by physiological and biochemical tests as B. subtilis strain type35,45 (Table 1). Nevertheless, the strains showed some differences between them, the A-SRETCR and M-SRETCR strains grew favorably at pH 5.0, 7.0, and 9.0 and at 2.0%, 5.0% and 7.0% NaCl, whereas the N-SRETCR strain grew at pH 5.0 and 7.0 and only at 2.0% NaCl. All three strains were rod-shaped when cultured in plates with CMC at pH 7.0 (Figs. 2A, C and D). The A-SRETCR strain also showed a filamentous morphology (Fig. 2B) in solid media at pH 5.0 and 9.0 and a pellet-shaped growth in liquid media at pH 5.0 and 9.0. Earl et al.15 reported that B subtilis grew in a chain-like fashion when the spores germinated under reduced carbon feed conditions. However, Mo and Burkholder33 showed that B. subtilis had filamentous growth when the YneA protein (a cell division inhibitor) was overexpressed as a response to environmental stress and DNA damage.
Morphological, physiological and biochemical characteristics of the strains isolated from composting with coffee residues and B. subtilis strain type.
| Characteristics | B. subtilis A-SRETCR | B. subtilis M-SRETCR | B. subtilis N-SRETCR | B. subtilise |
|---|---|---|---|---|
| Cell morphology | Filamentous and Rod-shaped | Rod-shaped | Rod-shaped | Rod-shaped |
| Cell size (μm) | 2.23×0.89 | 1.65×0.63 | 2.34×0.66 | 2.0–3.0×0.7–0.8 |
| Motility | + | + | + | + |
| Gram reaction | + | + | + | + |
| Endospores | − | Central | Terminal | Central and subterminal |
| Exopolysaccharides | − | + | − | + |
| Colony morphology | Circular, median, white, powdery texturea | Irregular, big, bright red center, flatb | Irregular, median, black-bright red, convexc | Irregular with appearance of mixed culture |
| Liquid medium | Pellets | Thin layer on the surface of the liquid | Cloudy | Thin layer on the surface of the liquidd |
| Growth | ||||
| Temperature (°C) | 37 | 37 | 37 | 37 |
| pH | 5.0, 7.0, 9.0 | 5.0, 7.0, 9.0 | 5.0, 7.0 | 5.0, 7.0, 9.0 |
| NaCl (%) | 2.0, 5.0, 7.0 | 2.0, 5.0, 7.0 | 2.0 | 2.0, 5.0, 7.0 |
| Utilization of | ||||
| Citrate | − | − | − | + |
| Crystalline cellulose | + | + | + | + |
| Carboxymethylcellulose | + | + | + | + |
| Hydrolysis of | ||||
| Casein | + | + | + | + |
| Starch | + | + | − | + |
| Gelatin | − | − | − | + |
| Acid from | ||||
| D-Glucose | + | + | + | + |
| D-Mannitol | − | + | + | + |
| D-Xylose | + | + | + | + |
| Cellobiose | + | + | + | + |
| Urea | − | − | − | − |
| Catalase | + | + | − | + |
| Oxidase | − | − | − | ± |
| Methyl red | − | − | − | − |
| Voges-Proskauer | − | + | − | + |
| Nitrate reduction | − | + | + | + |
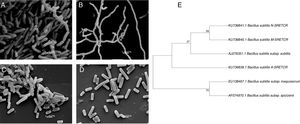
Scanning electron micrography of bacterial strains isolated from composting of coffee residues. (A) A-SRETCR cultured in medium with CMC pH 7.0; (B) A-SRETCR cultured in medium with CMC pH 5.0; (C) M-SRETCR and (D) N-SRETCR cultured in medium with CMC pH 7.0. (E) Phylogenetic analysis of 16S rDNA partial sequences of the strains of B. subtilis A-SRETCR, M-SRETCR and N-SRETCR isolated from composting of coffee residues and cow manure, using B. subtilis as reference sequences from bacterio.net LPSN. The phylogenetic tree was built with 1000 repetitions of bootstrap values by the method of maximum likelihood model Jukes-Cantor.
The analysis of partial 16S rDNA sequences revealed that the A-SRETCR and M-SRETCR strains have 99% similarity to B. subtilis subs. subtilis CU1050, whereas the N-SRETCR strain showed 99% similarity with B. subtilis I5-8. The identified strains were registered in GenBank under accession numbers KU736839.1 B. subtilis A-SRETCR, KU736840.1 B. subtilis M-SRETCR and KU736841.1 B. subtilis N-SRETCR. The phylogenetic analysis redundancy test (Fig. 2E) discarded the sequences of some B. subtilis reference sub-strains reported in the LPSN bacterio.net 2015 database but did not remove those of A-SRETCR, M-SRETCR and N-SRETCR, which suggested that although the three stains showed high similarity to B. subtilis, they were sufficiently different among each other and from the reference strains. Although they are related, B. subtilis variants are sub-classified into sub-species because they have some differences15. Likewise, Rooney et al.40 observed that B. subtilis sub-species are tightly related; their clear differentiation had not been possible based on phenotypic or phylogenetic characteristics and because the 16S rDNA is very conserved.
B. subtilis is known for its cellulolytic capacity and for its ability to develop in different environments14,15,37,42,55. In this study, B. subtilis strains A-SRETCR and M-SRETCR were isolated from composting piles with coffee residues in the thermophilic stage (61°C) at pH 9.3, whereas strain N-SRETCR was isolated from a composting pile at 57°C and pH 5.5. Acharya et al.2 reported the thermophilic strain of B. subtilis as having greater cellulolytic potential based on the diameter of the hydrolysis halos.
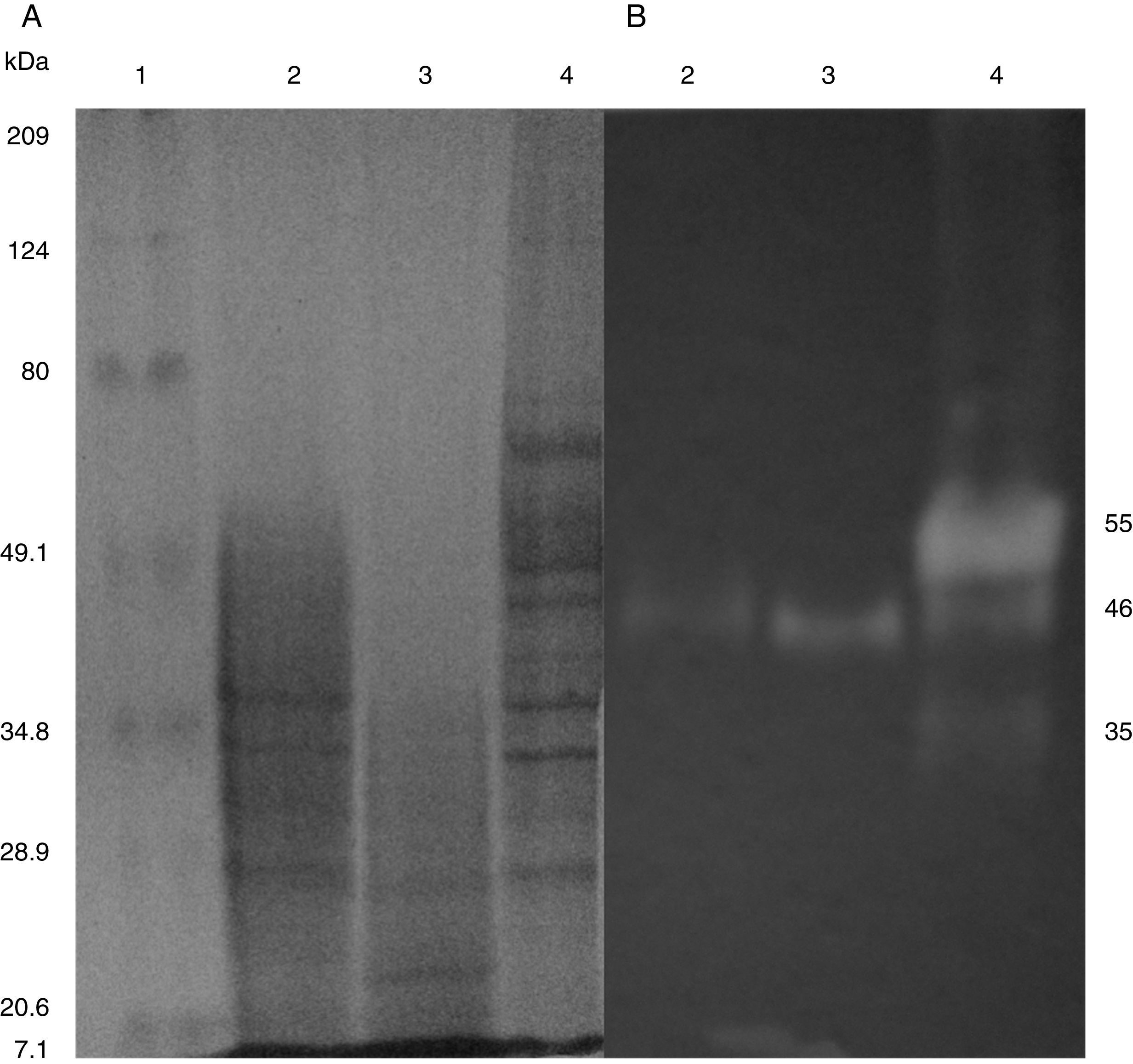
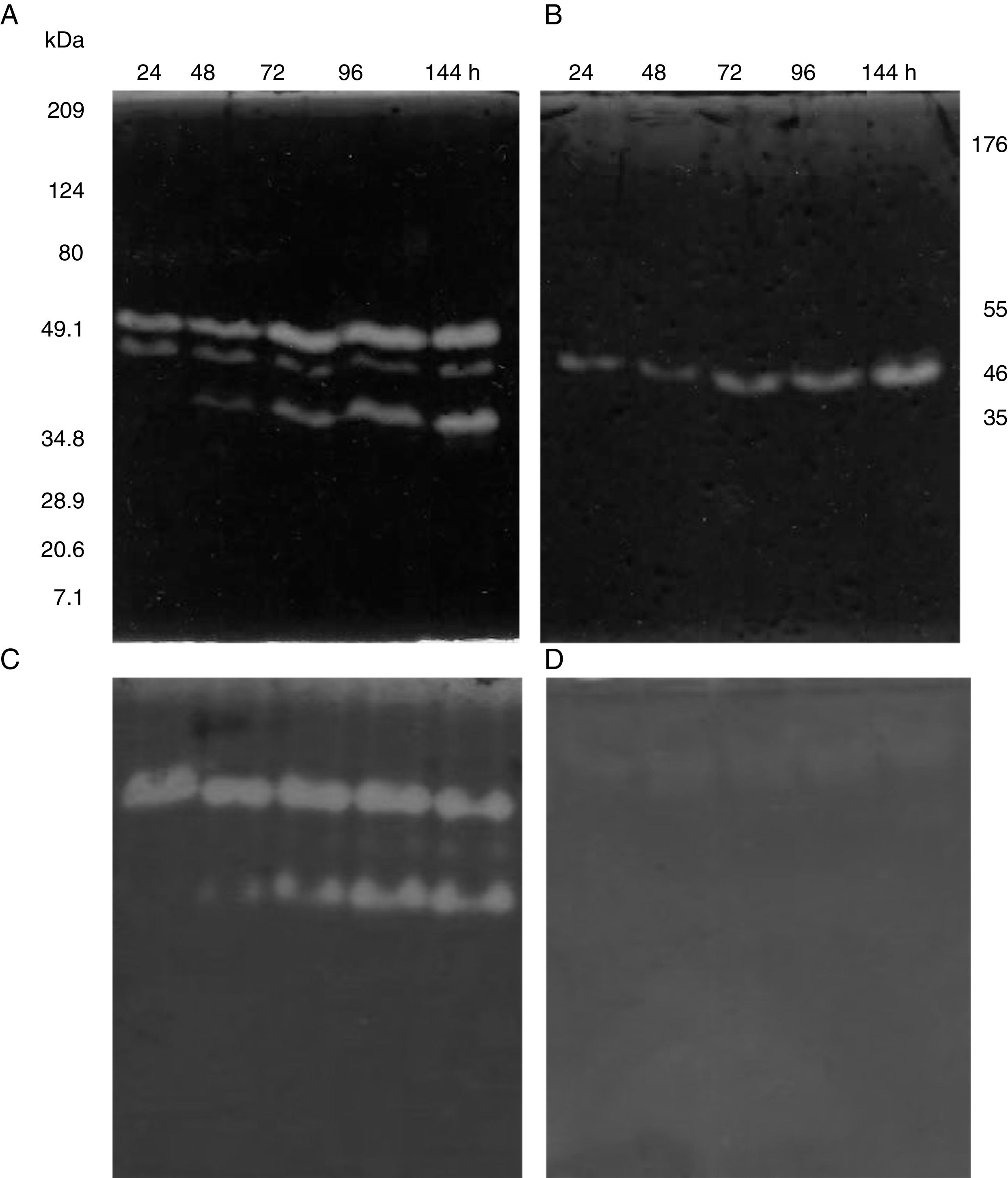
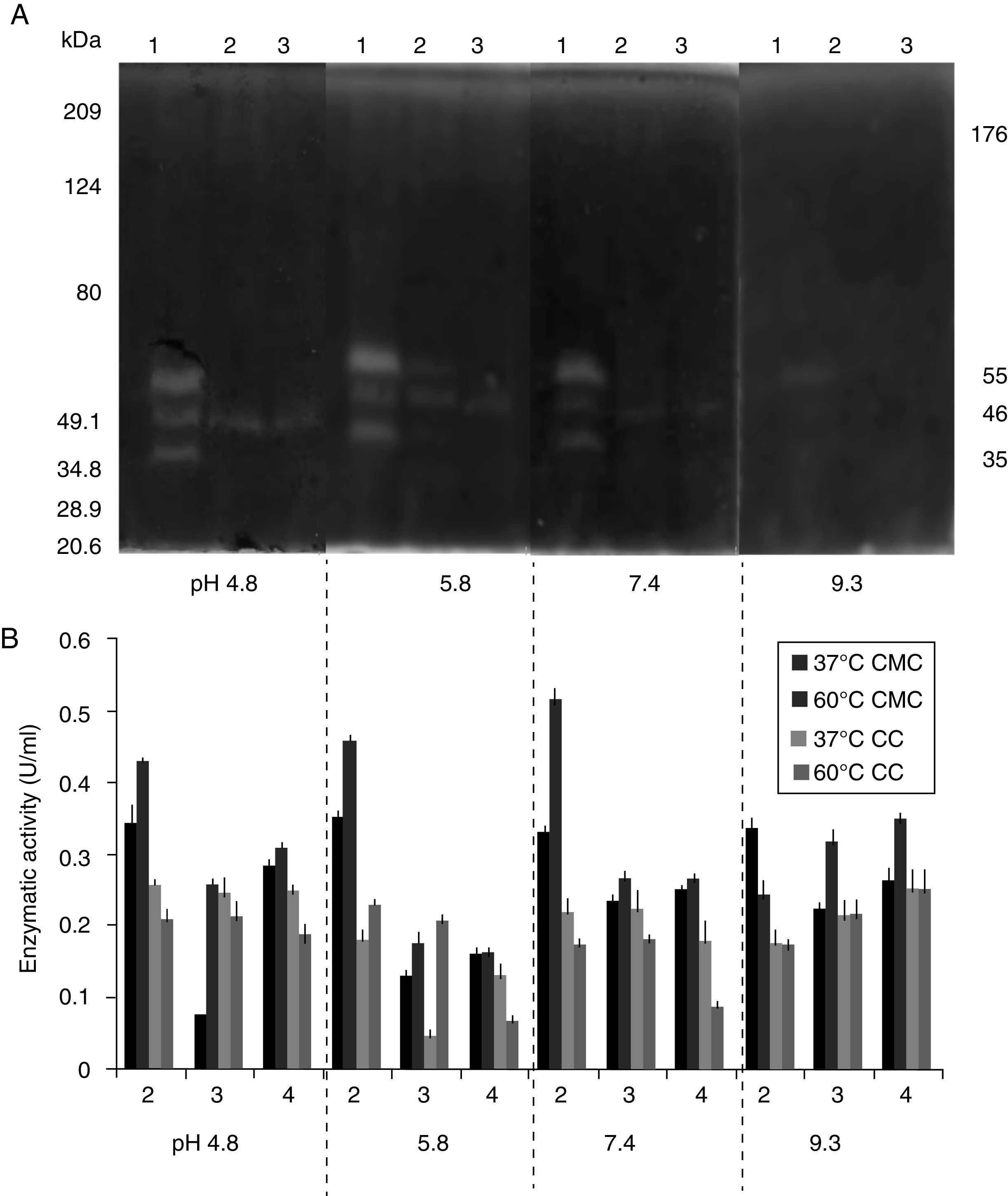
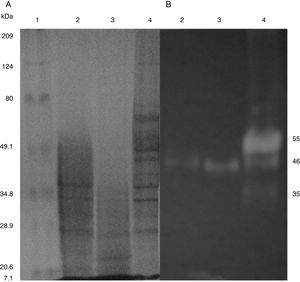
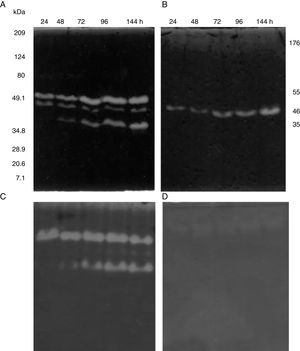
SDS-PAGE, Native-PAGE and zymogram analysesIn the profile of total proteins precipitated using thermal shock and separated by SDS-PAGE (Fig. 3A), the differences in the proteins secreted by all three strains can be observed. Their zymograms showed endocellulase activity (Fig. 3B) at pH 7.4 and 25°C. The intensity of the bands showed that the cellulase secreted by B. subtilis A-SRETCR showed higher activity. For all three strains, the zymograms revealed an endocellulase with an approximate molecular weight of 46kDa (Fig. 3B); A-SRETCR showed a hydrolysis band of 55kDa, similar to the one reported for different B. subtilis strains of 55 kDa23,28 and 56kDa11. Additionally, after 24h (Fig. 4), B. subtilis A-SRETCR showed hydrolysis bands corresponding to 35kDa, and M-SRETCR showed one band at 176kDa. The same number of bands was observed in SDS-PAGE and Native-PAGE, confirming that the extracts contained at least two endocellulases.
(A) SDS-PAGE of protein extract from a 24h culture in medium CMCYE pH 5.95, incubated at 37°C and 200rpm, precipitated with thermal shock and stained with Coomassie blue. (B) Zymogram of protein extract, incubated with 1× PBS, pH 7.4 and stained with 0.5% Congo red. Lane 1: Marker Prestained SDS-PAGE Broad range; lane 2: N-SRETCR; lane 3: M-SRETCR; and lane 4: A-SRETCR.
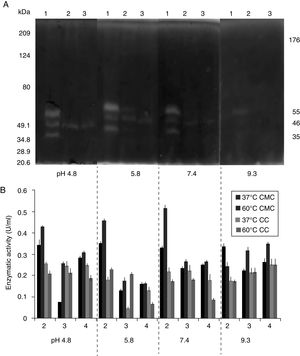
Several studies have reported single bands with endocellulase activity for different B. subtilis strains with maximum activities at pH 4–6 and 50°C and 60°C23,25,28,53–55. In this study, higher intensity bands were observed at acidic pH (4.8 and 5.8) and lower intensity bands were observed at pH 7.4 and 9.3 at 25°C (Fig. 5A) even after proteins were precipitated with drastic temperature changes (99°C and −20°C) and re-suspended with denaturing agents, such as SDS and 5.6M urea. This suggested that the cellulase extracts produced by B. subtilis A-SRETCR and M-SRETCR could have applications in different industries due to their activity at different pH and because they had recovered activity after being exposed to thermal shock. The use of cellulases at acidic pH has been suggested in the animal food industry and in the extraction and clarification of fruit juices9,31,47 as well as in the bleaching process of recycled paper2,3,50. Likewise, these cellulases could be used in the hydrolysis of coffee residues having an acidity of up to pH 4.5.
(A) Zymogram of the cellulase from a 24h culture in medium CMCYE pH 5.95, at 37°C and 200rpm. Protein was precipitated with thermal shock and separated in SDS-PAGE with 0.1% CMC, at 25°C, during 24h. (B) Enzymatic activity from protein extract in CMC and CC incubated to 37 and 60°C. Both were incubated at different pH values (0.1M citrate buffer at pH 4.8, 0.2M acetate buffer at pH 5.8, 1× PBS at pH 7.4 and 0.025M borate buffer at pH 9.3). Lanes 1: A-SRETCR; lane 2: M-SRETCR; lane 3: N-SRETCR.
The protein extracts of all three strains showed cellulolytic activity on CMC and CC (Fig. 5B), suggesting the presence of endo and exocellulases because the activity of these enzymes is specific for the presence of these substrates27,43. Although, exocellulase activity was detected in all extracts, higher activity (0.519U/ml) was observed on CMC at 60°C, which was similar to that reported for cellulases purified and characterized from B. subtilis with enzymatic activities between 0.200 and 0.600U/ml23,25,28,53–55. The extract of B. subtilis A-SRETCR had the highest activity on CMC at 37°C and 60°C for all four evaluated pH values. However, the activity was higher with PBS, which is a saline buffer containing Na+ and K+ ions. These constituents could have acted as cofactors and increased the enzymatic activity. Rawat and Tewari37 and Zhu et al.55 showed increases in cellulase activity in the presence of these ions.
The highest exocellulase activity (0.254U/ml) was observed in the extract of B. subtilis N-SRETCR at pH 9.3 and both temperatures and at pH 4.8 and 37°C, which could become the standard for future studies because no reports on exocellulases have identified optimal activity at this pH. Deka et al.14 reported an endocellulase of B. subtilis AS3 with optimal activity at pH 9.2 and suggested its use for the production of alternative fuels, such as bioethanol.
The isolation of microorganisms from composting piles in the thermophilic stage (61°C) allowed the identification of three B. subtilis strains whose enzymatic extracts showed activity on CMC and crystal cellulose, confirming the presence of endo and exocellulases. The cellulases of these microorganisms possessed cellulolytic activity at acidic pH (4.8 and 5.8). These cellulases could be used in several industries, such as in the preparation of animal foods, fruit juice extraction and clarification, and bleaching recycled paper. Moreover, cellulolytic activities were also observed at basic pH (9.3). These cellulases could be used in the cellulose and paper industries and in detergents, where enzymes able to tolerate basic pH are needed. However, it was observed that the cellulases of B. subtilis A-SRETCR had higher activity at higher temperatures (60°C). The activity recovery of the extracts with cellulolytic activity after exposure to drastic temperature conditions and chaotropic agents indicated their potential applications in industry, where typical operation conditions are known to cause enzyme denaturation due to mechanical damage and pH or temperature changes. Finally, the cellulolytic enzymes found in these microorganisms isolated from thermophilic conditions and from low acidity coffee processing residues could be used to take advantage of lignocellulosic residues generated in wet coffee processing. In turn, these complex residues could be used as fermentable substrates for the production of bioethanol.
Ethical disclosuresProtection of human and animal subjectsThe authors declare that no experiments were performed on humans or animals for this study.
Confidentiality of dataThe authors declare that no patient data appear in this article.
Right to privacy and informed consentThe authors declare that no patient data appear in this article.
Conflict of interestThe authors declare that they have no conflicts of interest.
This work was funded by ciencia-básica from Consejo Nacional de Ciencia y Tecnología (CONACyT) number: 180501 and El Colegio de la Frontera Sur, research center from Chiapas, México. HYSR, gratefully acknowledges the scholarship from CONACyT.