In patients with invasive fungal infections, the accurate and rapid identification of the genus Candida is of utmost importance since antimycotic sensitivity is closely related to the species. The aim of the present study was to compare the identification results of species of the genus Candida obtained by BD Phoenix™ (Becton Dickinson [BD]) and Maldi-TOF MS (Bruker Microflex LT Biotyper 3.1). A total of 192 isolates from the strain collection belonging to the Mycology Network of the Autonomous City of Buenos Aires, Argentina, were analyzed. The observed concordance was 95%. Only 10 strains (5%) were not correctly identified by the BD Phoenix™ system. The average identification time with the Yeast ID panels was 8h 22min. The BD Phoenix™ system proved to be a simple, reliable and effective method for identifying the main species of the genus Candida.
En pacientes con infecciones fúngicas invasoras, la identificación certera y rápida de las especies del género Candida es de suma importancia, ya que la sensibilidad a los antifúngicos está íntimamente relacionada con la especie. El objetivo del presente estudio fue comparar los resultados de identificación de especies del género Candida obtenidos con el equipo comercial BD PhoenixTM (Becton Dickinson [BD]) y con la técnica de Maldi-TOF MS (Bruker Microflex LT Biotyper 3.1.) Se analizaron 192 aislamientos provenientes del cepario perteneciente a la Red de Micología de la Ciudad Autónoma de Buenos Aires, Argentina. La concordancia observada fue del 95%. Solo 10 cepas (5%) no fueron identificadas correctamente por el sistema BD PhoenixTM. El tiempo promedio de identificación con los paneles Yeast ID fue de 8 h 22 min. El sistema BD PhoenixTM demostró ser un método simple, confiable y efectivo para la identificación de las principales especies del género Candida.
Candida albicans continues to be the most frequent yeast isolated from clinical specimens. However, Candida species distribution has been changing during the last decades3,5,7 and it varies based on different factors including region, patient's age group, underlying disease, prophylaxis used2,7. Candida glabrata is an important emergent pathogen in Northern Europe, the United States and Canada, whereas Candida parapsilosis prevails in Southern Europe, Asia and South America5. In developed countries C. glabrata ranks second, followed by C. parapsilosis and Candida tropicalis3. In Latin America, C. parapsilosis is found in the second place, followed by C. tropicalis and C. glabrata7. In Argentina, C. albicans has been reported to be in the first place, followed by C. parapsilosis and then C. tropicalis and C. glabrata, in studies performed by the National Mycology Network [Instituto Nacional de Microbiología C. Malbrán – Administración Nacional de Laboratorios e Institutos de Salud] and the Buenos Aires City Mycology Network2,6 thus confirming the data reported for Latin America7.
Among other factors, species distribution varies depending on the different stages of the life course, as it can be observed at the extremes of the age spectrum; while C. albicans and C. parapsilosis prevail in newborns, C. glabrata predominates in patients over 60 years old2,7.
The importance of a definitive identification lies on the fact that certain Candida species can show or develop a reduced sensitivity to one or several antifungal drugs, thus leading to crucial clinical consequences4,12. A quick turnaround time for this test is critical in current clinical settings, since a rapid diagnosis would enable the prescription of a specific antifungal treatment. On the other hand, identification down to species level allows for a better understanding of the epidemiology of each healthcare facility, which is important when establishing an empirical therapy.
The conventional methodology is laborious and slow, therefore, commercial methods allowing a faster identification have been developed. These methods, such as API 20C AUX® and API ID 32C® (bioMérieux, France), and RapID™ Yeast Plus® (Remel)9,14, are based on sugar assimilation and fermentation1.
During the last decades, automated methods appropriate for microbiology laboratories have been developed. Examples of these are Microscan WalkAway RYID® (Rapid Yeast Identification), Vitek YBC® (Yeast Biochemical Card), Vitek 2 ID-YST® and BD Phoenix™ Yeast ID, which can have yeasts identified within 4–48h. The automated microbiology system BD Phoenix™ Yeast ID has been designed for the rapid identification of clinically relevant yeasts9. The assays used in the panels are modifications of standard biochemical methods. They include tests for fermentation, oxidation, degradation and hydrolysis of various substrates, as well as single-carbon-source substrates, both chromogenic and fluorogenic. Use and degradation of specific substrates are detected by different indicator systems. Acid production from carbohydrates is detected by a change in the phenol red indicator. Chromogenic substrates contain p-nitrophenyl or p-nitroanilide compounds that produce a yellow color once its enzymatic hydrolysis has taken place. Enzymatic hydrolysis of fluorogenic substrates results in the release of a product derived from the fluorescent substance coumarin. Organisms using a single-carbon-source reduce a resazurin-based indicator10,14.
The purpose of this study was to compare the identification results obtained for different species of the genus Candida with either the BD Phoenix™ Yeast ID automated method or the matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry (MS) method.
A collection of 192 strains of the genus Candida, coming from various clinical specimens processed at member hospitals of the Buenos Aires City Mycology Network, Argentina, between 2009 and 2014, was analyzed. All strains were identified by conventional methods which included: micromorphological study in corn-meal agar (Oxoid, UK) with 1% Tween 80; chromogenic agar for yeasts (CHROMagar™ Candida, Paris, France); API® ID 32C or API® 20C AUX (bioMérieux, Marcy l’Etoile, France) Biochemical tests were added to differentiate C. albicans from Candida dubliniensis, developmental capacity at different temperatures, production of chlamydoconidia in different substrates and production of halo of opacity in media containing Tween 80 and CaCl28.
Thirty-nine C. tropicalis, 34 Candida guilliermondii, 31 C. albicans, 29 C. parapsilosis, 28 C. glabrata, 23 Candidakrusei, 4 C. dubliniensis, 3 Candida lusitaniae and 1 Candida lipolytica were tested. The strains were inoculated onto chromogenic culture media for yeasts (CHROMagar™ Candida) in order to confirm purity. Each isolate was subcultured in Sabouraud dextrose agar and incubated for 48h, at 35°C, before using the BD Phoenix™ Yeast ID panels as instructed by the manufacturer9. As the Phoenix system does not report confidence values lower than 90%, results with scores >90% were considered to represent acceptable identifications.
Identification results were compared with those obtained by MALDI-TOF MS (Bruker Microflex LT Biotyper 3.1). The MALDI ionization, coupled to a TOF analyzer is a soft ionization technique used in mass spectrometry, which allows the analysis of biomolecules and large organic molecules11,13.
MALDI-TOF MS was used as reference method, together with genotyping identification in those strains yielding discrepant identification results. For the molecular technique, DNA extraction was performed, followed by amplification of specific DNA portions with universal primers ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) by PCR. Amplification products were sequenced using the Sanger method. The BLAST tool was used for identification and the criteria established were concordance >97% and coverage of at least 99%.
Candida krusei ATCC 6258, C. parapsilosis ATCC 22019 and C. albicans ATCC 64548 were used as control strains.
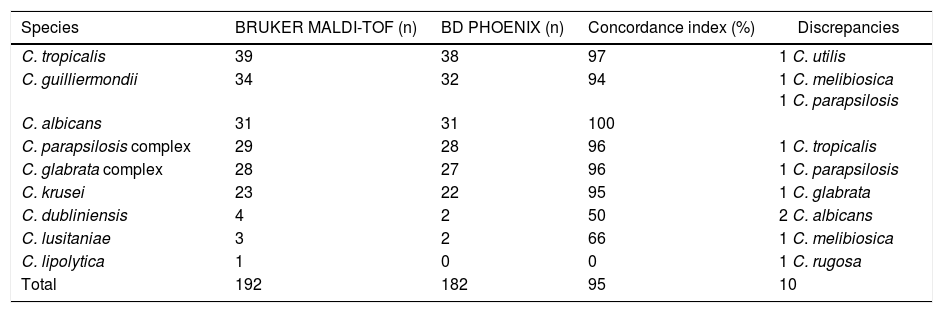
Among the different species studied, concordance was variable and rates are detailed in Table 1. The general concordance index was 95%. While C. albicans showed a concordance rate of 100%, the remaining species evidenced at least one discrepancy with the reference method. Ten strains (5%) were misidentified with the Yeast ID panels: one C. tropicalis, two C. guilliermondii, one C. parapsilosis, one C. krusei, two C. dubliniensis, one C. lusitaniae and one C. lipolytica. The low concordance rates obtained for the last three species mentioned above can be due to the low number of strains tested. For the remaining species studied the index was ≥96%.
Concordance index and discrepancies obtained when comparing the BD Phoenix™ system with the MALDI-TOF
| Species | BRUKER MALDI-TOF (n) | BD PHOENIX (n) | Concordance index (%) | Discrepancies |
|---|---|---|---|---|
| C. tropicalis | 39 | 38 | 97 | 1 C. utilis |
| C. guilliermondii | 34 | 32 | 94 | 1 C. melibiosica 1 C. parapsilosis |
| C. albicans | 31 | 31 | 100 | |
| C. parapsilosis complex | 29 | 28 | 96 | 1 C. tropicalis |
| C. glabrata complex | 28 | 27 | 96 | 1 C. parapsilosis |
| C. krusei | 23 | 22 | 95 | 1 C. glabrata |
| C. dubliniensis | 4 | 2 | 50 | 2 C. albicans |
| C. lusitaniae | 3 | 2 | 66 | 1 C. melibiosica |
| C. lipolytica | 1 | 0 | 0 | 1 C. rugosa |
| Total | 192 | 182 | 95 | 10 |
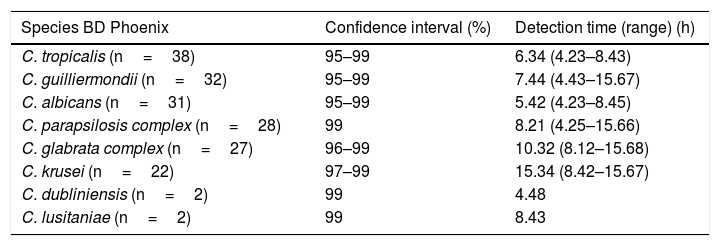
Detection turnaround times for each of the species are shown in Table 2, with a mean time of 8h 22min (range 4h 23min–15h 68min). MALDI-TOF MS differentiated two of the species of the C. parapsilosis complex as Candida orthopsilosis and one species of the C. glabrata complex as Candida bracarensis. The BD Phoenix™ system did not differentiate the cryptic species from those complexes, because it is a method that identifies microorganisms based on the phenotypic features of yeasts.
Detection time of each Candida species
| Species BD Phoenix | Confidence interval (%) | Detection time (range) (h) |
|---|---|---|
| C. tropicalis (n=38) | 95–99 | 6.34 (4.23–8.43) |
| C. guilliermondii (n=32) | 95–99 | 7.44 (4.43–15.67) |
| C. albicans (n=31) | 95–99 | 5.42 (4.23–8.45) |
| C. parapsilosis complex (n=28) | 99 | 8.21 (4.25–15.66) |
| C. glabrata complex (n=27) | 96–99 | 10.32 (8.12–15.68) |
| C. krusei (n=22) | 97–99 | 15.34 (8.42–15.67) |
| C. dubliniensis (n=2) | 99 | 4.48 |
| C. lusitaniae (n=2) | 99 | 8.43 |
h, hours.
According to the results obtained, we can conclude that the BD Phoenix™ Yeast ID panel system is a reliable, useful and rapid method for routine identification of the yeast species most frequently isolated from clinical specimens. Since mass spectrometry methods such as MALDI-TOF are not available in most clinical microbiology laboratories, the BD Phoenix™ system offers an effective alternative. It is noteworthy that identification was completed in only 8h 22min on average and with a concordance index of 95%.
Conflict of interestThe authors declare that they have no conflicts of interest.
We are very grateful to Dr. María Magdalena Peres from Becton Dickinson Argentina who generously provide us with Yeast-ID panels from the system BD Phoenix™.
Ana Maria Romeo (Htal. Penna), Mariela Schijman (Htal. Álvarez), Claudia Garbasz (Htal Pirovano), Graciela Ponce (IREP), Silvana Cataldi (Htal Durand), Laura Dufranc (Htal Zubizarreta), Nora Franco (Htal. Piñero), Mónica López (Htal Ramos Mejía), Ricardo Iachini (I. Pasteur), Rosana Pereda (Htal P. Elizalde), Alicia Arechavala (Hospital Muñiz), Analia Fernandez (F. Favaloro), Silvia Relloso (CEMIC), Ivana Maldonado (Htal Aleman), Agustina Forastiero (Htal Britanico), Norma Fernandez (Htal de Clinicas).