Entamoeba infections occur worldwide, with higher frequency in countries of low socioeconomic status and poor public health. Since Entamoeba histolytica has long been recognized as the only pathogenic species, making a differential diagnosis of other morphologically identical Entamoeba is important. This study aimed to determine the prevalence of Entamoeba species in two populations from Argentina, make a differential diagnosis by PCR and characterize Entamoeba isolates at the SSU rRNA gene. A total of 493 serial fecal samples were obtained from individuals in the provinces of Buenos Aires (n=210) and Misiones (n=283). Samples were examined by conventional methods (formalin–ethyl acetate and Willis flotation) and specific PCRs to differentiate Entamoeba species. Entamoeba isolates were characterized by sequencing a fragment of the SSU rRNA gene. The overall prevalence of Entamoeba infection was 12.4%, being more prevalent in Buenos Aires than in Misiones (14.8% vs. 10.6%). A case of E. histolytica confirmed by PCR and sequence analysis was reported for the first time in Buenos Aires. Moreover, new genetic data on Entamoeba coli and Entamoeba dispar were recorded. The phylogenetic analysis revealed a congruence between morphological characteristics and SSU rRNA gene sequences. This study increases the amount of information on the distribution of these species in Argentina and the region of the Americas.
Las infecciones por Entamoeba se producen en todo el mundo, con mayor frecuencia en países de bajo nivel socioeconómico y salud pública deficiente. Dado que se ha reconocido a Entamoeba histolytica como la única especie patógena del género, es importante realizar un diagnóstico diferencial respecto de otras especies de Entamoeba morfológicamente idénticas. Este estudio tuvo como objetivos determinar la prevalencia de especies de Entamoeba en 2 poblaciones de Argentina, realizar su diagnóstico diferencial por PCR y caracterizar los aislados de Entamoeba secuenciando un fragmento del gen SSU ARNr. Se obtuvieron 493 muestras fecales seriadas de individuos de las provincias de Buenos Aires (n=210) y Misiones (n=283). Las muestras se examinaron por métodos convencionales (sedimentación de formalina-etil acetato y flotación de Willis) y mediante PCR específicas para diferenciar especies de Entamoeba. Los aislamientos de Entamoeba se caracterizaron por secuenciar un fragmento del gen SSU ARNr. La prevalencia general de la infección por Entamoeba fue del 12,4% y fue mayor en Buenos Aires que en Misiones (14,8 vs. 10,6%). Se informó por primera vez un caso de Entamoeba histolytica en Buenos Aires, confirmado por PCR y análisis de secuencia. Además, se registraron nuevos datos genéticos sobre Entamoeba coli y Entamoeba dispar. El análisis filogenético reveló una congruencia entre las características morfológicas y las secuencias del gen SSU ARNr. A través de este estudio, hemos sumado información acerca de la distribución de estas especies en nuestro país y en la región de las Américas.
The protozoan species Entamoeba dispar, Entamoeba histolytica, Entamoeba moshkovskii, and Entamoebabangladeshi are collectively referred to as the Entamoeba complex because they are morphologically identical1. Among these species, E. histolytica is the causative agent of amebiasis in humans, which is associated with intestinal and/or extra-intestinal manifestations. Infection typically initiates with the ingestion of mature cysts found in food, water, or hands contaminated with stool5,21. Although amebiasis is recognized as a neglected emerging disease, most cases are asymptomatic. Individuals with symptomatic amebiasis often suffer from amebic colitis and amebic liver abscess24. Symptoms of amebic colitis can range from mild diarrhea to severe dysentery, with abdominal pain and watery or bloody diarrhea18. Meanwhile, severe chronic infections may lead to further complications such as peritonitis, perforations, and the formation of amebic granulomas21.
On the other hand, the pathogenicity of E. moshkovskii and E. dispar remains unclear7. Recent studies have shown that E. moshkovskii can cause gastrointestinal disorders13,33. Furthermore, previous studies have indicated an association between E. dispar and clinical symptoms19,41. Meanwhile, E. bangladeshi has been isolated from fecal samples of both asymptomatic children and those experiencing diarrhea14.
Traditionally, amebic infections have been diagnosed by microscopy; however, this method relies on the observers’ experience and cannot distinguish among the Entamoeba complex species. On the other hand, many diagnostic tools based on stool polymerase chain reaction (PCR) have the highest sensitivity in distinguishing species34. In this sense, the small subunit rRNA (SSU rDNA) gene has been widely used to analyze phylogenetic relationships among eukaryotic organisms and, mainly, to detect Entamoeba species in stool samples36. Entamoeba SSU rRNA can be found in multiple copies of extrachromosomal plasmids and is relatively fast-evolving; hence, providing sufficient resolution to differentiate Entamoeba taxa12.
Non-pathogenic amebae, such as Entamoeba coli, Entamoeba polecki, Entamoeba hartamanni, Endolimax nana, and Iodamoeba bütschlii may also be confused with E. histolytica in diagnostic investigations5. Therefore, an accurate diagnostic strategy is needed to avoid misdiagnosis and wrong treatment24.
In the region of the Americas, E. coli was determined as the most prevalent and widely distributed ameba by conventional methods (e.g., direct smear and formalin–ethyl concentration), followed by the Entamoeba complex. However, more studies based on molecular techniques are needed to corroborate if conventional methods overestimate the prevalence31.
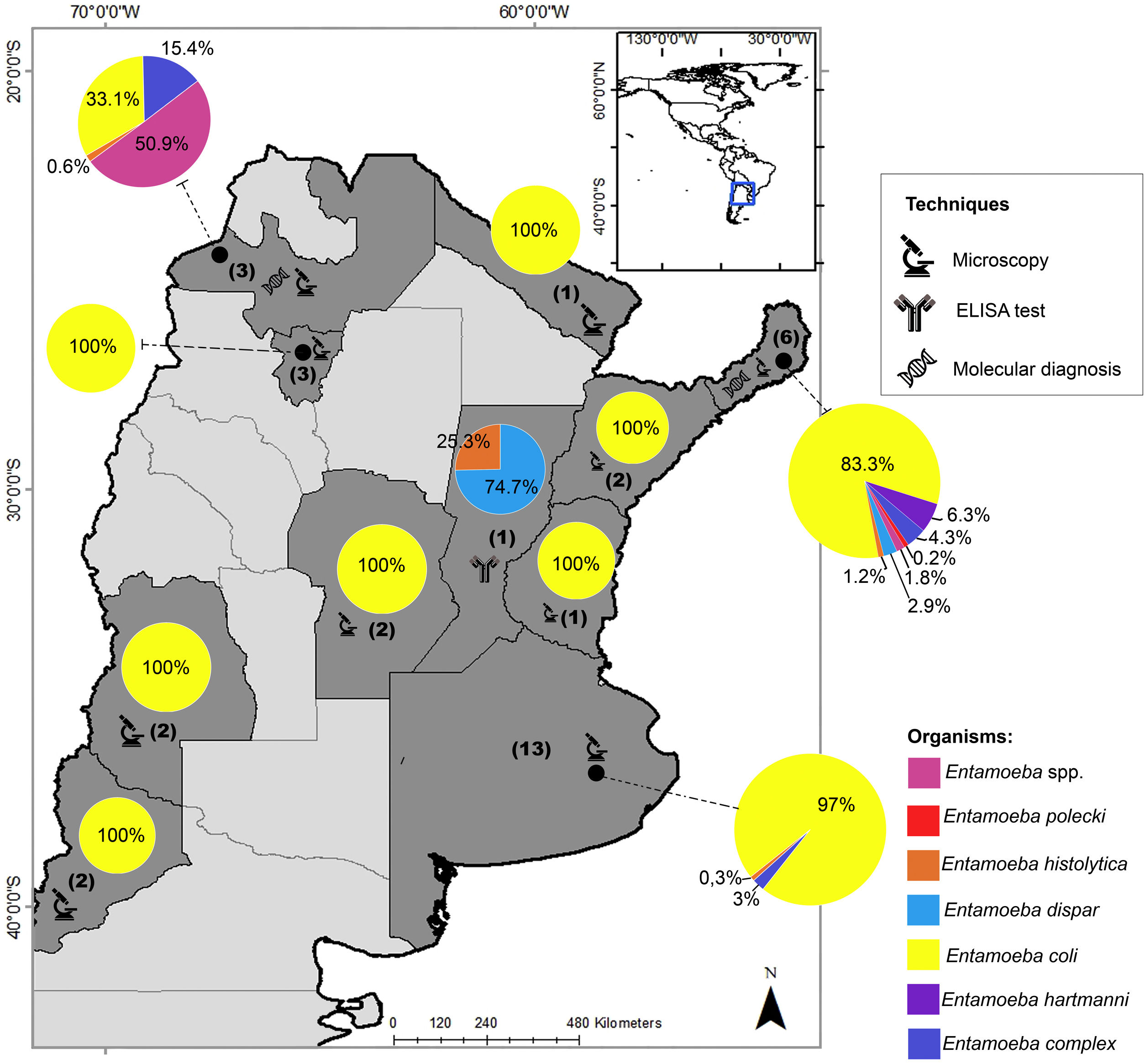
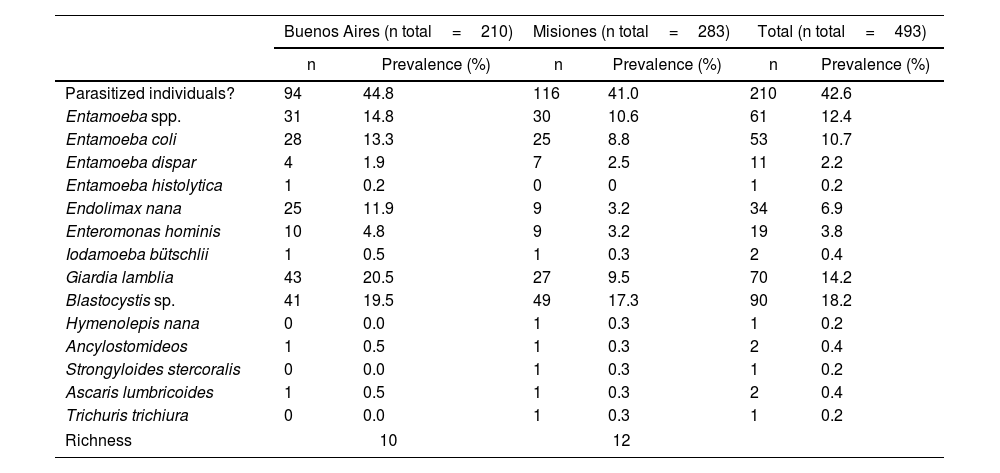
In Argentina, there are limited studies regarding the differential prevalence of Entamoeba species. In the last three decades, most of the prevalence studies were mainly based on microscopy and reported E. coli as the most frequent species (Fig. 1). In addition, most of these studies reported the diagnosis as Entamoeba complex, which was usually determined by microscopy20. Only one research performed an ELISA test and found a seroprevalence of 28% (21/75) of E. histolytica in a native community from the province of Santa Fe2. This protozoan parasite was also detected in Buenos Aires by microscopic examination with a low prevalence (0.9%; 2/210)9. E. histolytica was detected by molecular methods in 1 out of 99 individuals analyzed in Salta8 and detected in 10 out of 218 individuals studied in Misiones4. In the latter province, different studies detected up to six species of Entamoeba infecting humans, mainly by morphological diagnosis4,23,29,42,43 (Fig. 1).
Geographical distribution of Entamoeba spp. detected by studies performed in Argentina in the last three decades. Pie charts represent the proportion in which each Entamoeba species was detected. The number of studies that examined the prevalence of amebae in each province of Argentina is shown in brackets. Techniques performed for diagnosis are also represented. The map was drawn up based on Supplementary Table 1. Maps were performed using QGIS version 3.12 (Quantum GIS Development Team, 2020).
In light of scarce molecular data on Entamoeba species in Argentina, the present study aimed to: (i) determine the prevalence of Entamoeba species in human fecal samples from two populations of this country by microscopic methods, (ii) make a differential diagnosis by PCR and (iii) characterize the Entamoeba isolates at the SSU rRNA gene.
Material and methodsStudy area and sample collectionData for this study were obtained as part of a larger cross-sectional survey on intestinal parasitic infections and socioenvironmental conditions in human populations, which was conducted in schools, primary health care centers, and communal dining rooms located in two Argentine provinces: Buenos Aires and Misiones, between 2018 and 2019.
In Buenos Aires, 210 fecal samples were obtained from individuals aged 1–64 years residing in peripherical areas of the district of La Plata (34°55′17.22″S 57°57′16.31″W) and three municipalities in the South of Greater Buenos Aires (34°42′39.30″S 58°21′26.65″W; 34°42′33.45″S 58°18′17.22″W; 34°49′17.59″S 58°20′45.99″W). These individuals were characterized as living in poor socioeconomic conditions, including no piped water connection, cesspit system, and informal works.
In Misiones, 283 fecal samples were obtained from individuals aged 1–64 years residing in metropolitan areas surrounding the locality of Aristóbulo del Valle (27°05′43″S 54°53′49″W). These individuals lived in houses made of sheet metal and wood, most of whom consumed water from wells and had no access to the sewage system.
After informed consent was obtained, each individual was provided with a sterile plastic vial containing 70% ethanol. Stool samples from 3, consecutive or intermittent days, were collected from each participant.
When participants were children, samples were collected by their parents or legal guardians.
Fecal samples were submitted to the Centro de Estudios Parasitológicos y de Vectores (CEPAVE-CONICET-UNLP, La Plata, Buenos Aires, Argentina). The samples were divided into equal portions for microscopic examination and PCR sequencing analysis to differentiate all Entamoeba species.
Microscopic identification of Entamoeba cystsFecal samples were examined by light microscopy for the presence of Entamoeba and other intestinal parasites. The formalin–ethyl acetate concentration technique (FECT) and Willis flotation method were performed to detect the presence of parasites. First, each fecal sample, preserved in 70% ethanol, was homogenized with a ceramic pestle and mortar. The suspension was strained into a 15ml conical tube through a double layer of gauze placed into a strainer or small funnel and centrifuged at 1500rpm for 5min. The supernatant was discarded. Then, 7ml of formalin and 3ml of ethyl acetate were added to the sediment, mixed, and centrifuged at 1500rpm for 5min40. For Willis flotation, a saturated sodium chloride solution (S.G.=1.2g/ml) was used.
Molecular characterization of Entamoeba species in stool samplesDNA was extracted from fecal samples which were found to be positive for Entamoeba cysts. Before extraction, 500mg of feces/ethanol suspension was centrifuged at 8000g, and the pellet was washed three times with 1ml of Dulbecco's phosphate-buffered saline (PBS). After centrifugation, the pellet was subjected to mechanical disruption by three freeze–thaw cycles. DNA was isolated using the ZR Fecal DNA MiniPrep™ Kit (Zymo Research, California, USA) following the manufacturer's instructions32.
Entamoeba-specific PCRThe DNA obtained was used for amplification of a 135-bp fragment harboring the SSU rRNA gene by PCR with the primers ED1 (5′-TACAAAGTGGCCAATTTATGTAAGTA-3′), EH1 (5′-GTACAAAATGGCCAATTCATTCAATG-3′) for E. dispar and E. histolytica detection, respectively, with the unique reverse primer EHD2 (5′-ACTACCAACTGATTGATAGATCAG-3′) following the cycling conditions reported by Gonin and Trudel15.
DNA from samples identified as E. histolytica–E. dispar complex by the microscopic diagnosis, but testing negative for the above PCR, were amplified using the primers Entam1 (5′-CACTATTGGAGCTGGAATTAC-3′) and Entam2 (5′-GTTGATCCTAGTATTATATG-3′)39. Cycling conditions were: 5min at 95°C, 35 cycles of 30s at 95°C, 30s at 55°C and 30s at 72°C, with a final extension at 72°C for 2min.
In both cases, PCR conditions were optimized in a final volume of 25μl as follows: 1× GoTaq® buffer (Promega, Madison, USA), 0.2μM dNTPs, 1U/μl GoTaq® Hot Start polymerase (Promega, Madison, USA), 0.5μM of each primer, 0.1μg/μl BSA, 1.5mM MgCl2 and 4μl of DNA as template. Positive DNA samples were obtained from E. histolytica and E. dispar human fecal samples provided by the Division of Parasitic Diseases and Malaria (Centers for Disease Control and Prevention).
PCR products were further purified and sequenced (Macrogen, Seoul, Korea), along with both primers used for each amplification, to validate positive PCR results and perform phylogenetic analyses. Forward and reverse sequences obtained were assembled using the PREGAP and Gap4 programs of the Staden package35. The consensus sequences were compared to the previously published sequences of the Entamoeba genus by using the Basic Local Alignment Search Tool (BLAST) (www.ncbi.nlm.nih.gov/blast). Sequences obtained were deposited at GenBank under accession numbers: MZ787759–MZ787761, MK541024–MK541027, OM985615–OM985620 and ON712729.
Phylogenetic analysisThe phylogenetic analysis was carried out using newly obtained Entamoeba sequences and matching them with other representative sequences. Furthermore, the available GenBank sequence of the Endolimax nana (accession no. AF149916) was used as an outgroup (Supplementary Table 2). DNA sequence data were aligned using the CLUSTALW program6, with default options for introducing gaps into the alignment. When needed, inconsistencies were checked and manually edited. Phylogenetic relationships were inferred by maximum likelihood (ML) using MEGA X software22 and the Bayesian inference (BI) as implemented in MrBayes Ver. 3.2.6 software30. The best DNA substitution model (T92+G) was estimated using the jModelTest program26. Nodal support of ML analysis was estimated by performing 1000 bootstrap replications. The BI analysis was performed using Markov chain Monte Carlo (MCMC) chains for 1000000 generations with sample frequency set at 100. The first 25% of the trees sampled were discarded as ‘burn-in’. This number of generations was considered sufficient because the SD dropped below 0.01. A pairwise distance matrix among Entamoeba sequences was calculated using the nucleotide p-distance algorithm implemented in MegaX22.
Statistical analysesThe prevalence of parasitized individuals and parasitic species was calculated. A sample was considered positive when at least a parasitic species was detected by any morphological or molecular method. The prevalence was calculated as the number of parasitized individuals divided by the total number of individuals analyzed, expressed in terms of percentage.
The relationships among parasitism and infection with Entamoeba species and intestinal symptoms were analyzed statistically. The Chi-squared test (χ2) was used to determine the independence between the variables. In all cases, a p-value of <.05 was taken to indicate significance. The odds ratio (OR) and 95% confidence interval (95% CI) were calculated.
All statistical analyses were performed using R software version 4.2.128.
Ethics approvalThis study was approved by the Bioethics Consultative Committee of the National University of La Plata (Exp. No. 100–20 120/18) and the Research Ethics Committee Hospital El Cruce (Exp. No. 2919/1799/18). This research was conducted without affecting the physical, psychological or moral integrity of the participants, and protecting their identity, complying with the ethical standards established by the Nüremberg Code (1947), the principles proclaimed in the Universal Declaration of Human Rights (1948), the Declaration of Helsinki (1964) and its successive amendments. Special attention was also paid to Article 5 of the Regulation Decree of National Law 25.326. In addition, all families involved were given the results of the parasitological diagnosis, and positive cases were referred to the nearest health center to receive the corresponding antiparasitic treatment.
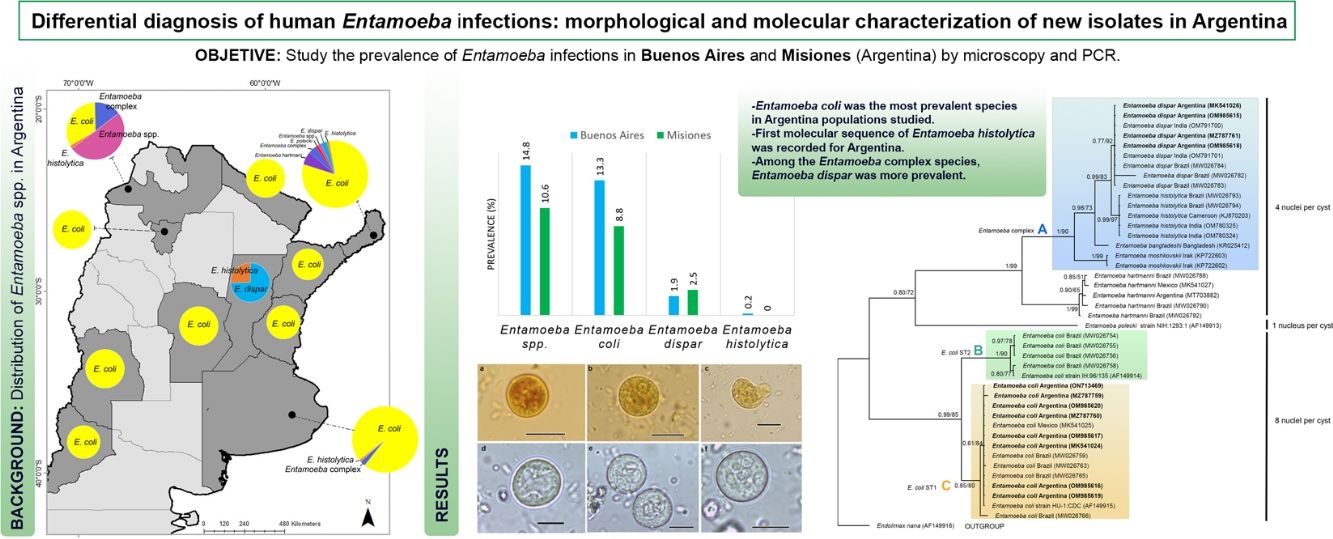
ResultsEntamoeba spp. prevalenceA total of 493 individuals aged 1–64 years were examined for Entamoeba cysts. The microscopic examination showed Entamoeba spp. in 12.4% (61/493) of the samples (Fig. 2). Among these, 86.9% (53/61) were identified as E. coli and 18.0% (11/61) as Entamoeba complex. By using PCR, E. dispar was recognized in 10 (2.0%), and E. histolytica in one (0.2%) of the total samples (Table 1) (Supplementary Fig. 1). The prevalence of these species and other intestinal parasites found in the analyzed provinces are shown in Table 1.
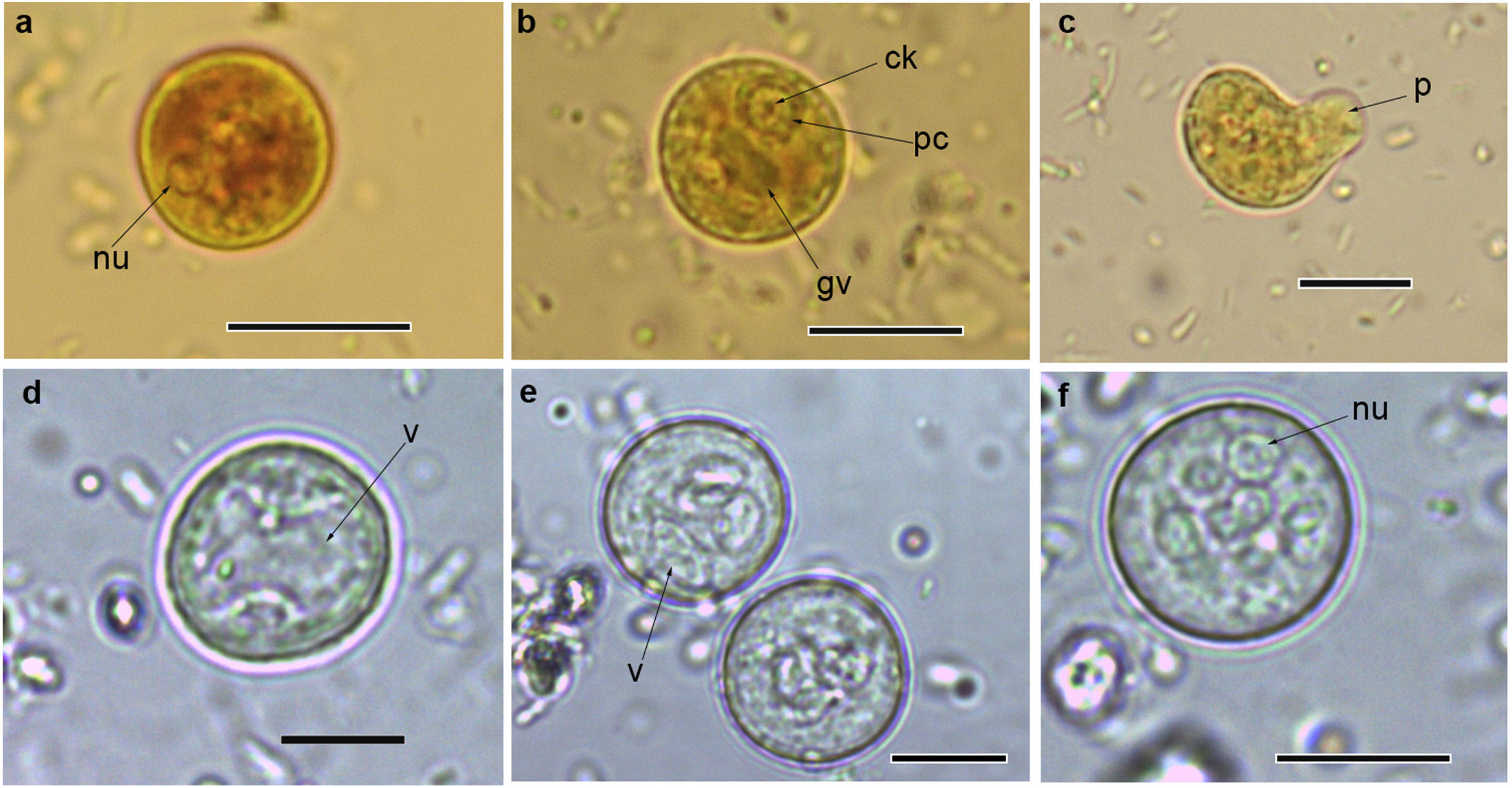
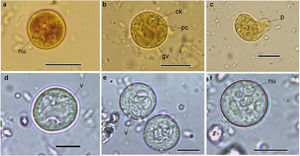
Cysts and trophozoites of Entamoeba found in populations from Argentina: (a and b) immature cysts of Entamoeba complex staining with Lugol; (c) trophozoite of Entamoeba complex staining with Lugol; (d and e) immature cysts of E. coli; (f) mature cyst of E. coli.Abbreviations: nu: nucleus; ck: central karyosome; gv: glycogen vacuole; pc: peripheral chromatin; v: vacuole; p: pseudopodia. Scale bars: a–f=10μm.
Prevalence of Entamoeba infection based on microscopy and PCR assay of human fecal samples according to Argentinian locations. Prevalence was calculated according to the total number of individuals analyzed in each province: 210 in Buenos Aires and 283 in Misiones. The prevalence of other intestinal parasites was also included. At the end of the table, parasite richness was indicated for each province.
| Buenos Aires (n total=210) | Misiones (n total=283) | Total (n total=493) | ||||
|---|---|---|---|---|---|---|
| n | Prevalence (%) | n | Prevalence (%) | n | Prevalence (%) | |
| Parasitized individuals? | 94 | 44.8 | 116 | 41.0 | 210 | 42.6 |
| Entamoeba spp. | 31 | 14.8 | 30 | 10.6 | 61 | 12.4 |
| Entamoeba coli | 28 | 13.3 | 25 | 8.8 | 53 | 10.7 |
| Entamoeba dispar | 4 | 1.9 | 7 | 2.5 | 11 | 2.2 |
| Entamoeba histolytica | 1 | 0.2 | 0 | 0 | 1 | 0.2 |
| Endolimax nana | 25 | 11.9 | 9 | 3.2 | 34 | 6.9 |
| Enteromonas hominis | 10 | 4.8 | 9 | 3.2 | 19 | 3.8 |
| Iodamoeba bütschlii | 1 | 0.5 | 1 | 0.3 | 2 | 0.4 |
| Giardia lamblia | 43 | 20.5 | 27 | 9.5 | 70 | 14.2 |
| Blastocystis sp. | 41 | 19.5 | 49 | 17.3 | 90 | 18.2 |
| Hymenolepis nana | 0 | 0.0 | 1 | 0.3 | 1 | 0.2 |
| Ancylostomideos | 1 | 0.5 | 1 | 0.3 | 2 | 0.4 |
| Strongyloides stercoralis | 0 | 0.0 | 1 | 0.3 | 1 | 0.2 |
| Ascaris lumbricoides | 1 | 0.5 | 1 | 0.3 | 2 | 0.4 |
| Trichuris trichiura | 0 | 0.0 | 1 | 0.3 | 1 | 0.2 |
| Richness | 10 | 12 | ||||
Entamoeba infection was more prevalent in the population from Buenos Aires (14.8%: 31/210), registering the first sequence data of E. histolytica in this province. In Misiones, the prevalence of Entamoeba spp. was 10.6% (30/283) (Table 1).
E. coli was the most prevalent species in both populations, followed by E. dispar (Table 1).
Statistically significant associations were observed between E. coli–E. nana (χ2=9.42; p=0.01; OR=3.40 [1.49–7.74]).
Molecular characterization of isolates and phylogenetic analysisEight PCR amplicons (467–631bp) were successfully obtained from isolates of E. coli (GenBank Acc. Nos. MK541024, MZ787759–60, OM985616–17, OM985619–20, ON713469). The alignment of these sequences showed 99.3% identity with four polymorphic sites. Pairwise genetic distance ranged from 0 to 0.58.
Four sequences (491–591bp) of E. dispar (MK541026, MZ787761, OM985615, OM985618) were 100% identical. In addition, one sequence of E. histolytica (133bp) (ON712729) was obtained.
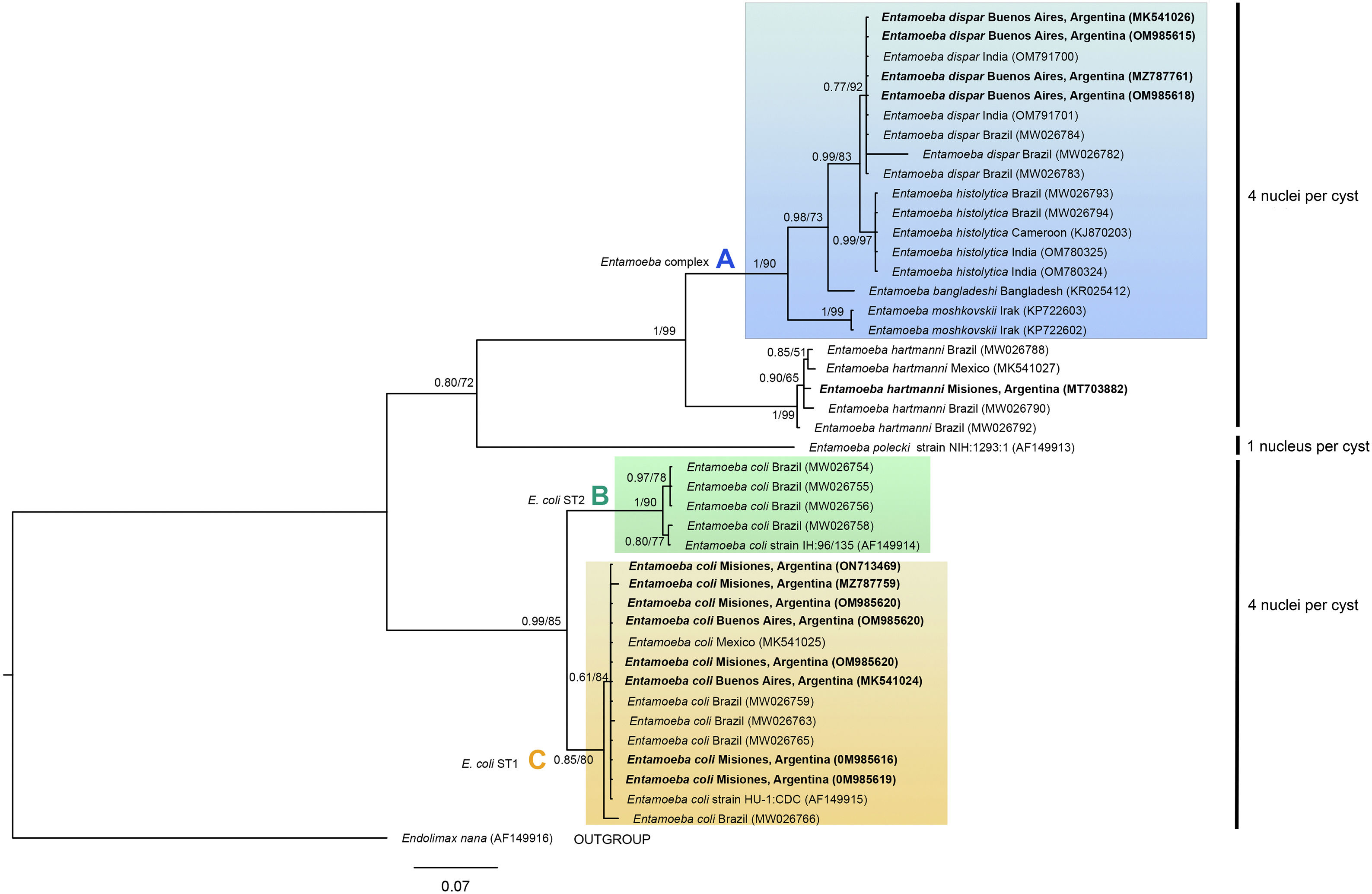
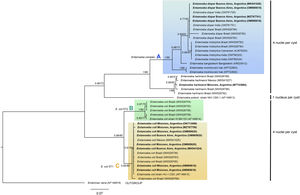
The phylogenetic reconstruction based on the ML and BI analyses showed the same topology. Three high-supported clades were formed, which correlated with species of Entamoeba-producing cysts with the same number of nuclei (Fig. 3). First, sequences of E. histolytica, E. dispar, E. moshkovskii, and E. bangladeshi isolated from humans clustered together (clade A). Sequences of Entamoeba hartmanni were placed as a sister lineage to clade A. All together formed a strongly supported monophyletic group including all species with four nuclei per mature cyst. The isolate of E. polecki, which forms cysts containing a single nucleus, appeared as a sister clade.
Phylogenetic tree constructed by the BI and ML methods based on 42 sequences of a partial region of the SSU rRNA gene under the substitution model HKY+G+I. Sequences are identified by taxon name and country of origin (when this data was available) and GenBank Acc. Nos. Sequences obtained in the present study are in bold. The posterior probabilities and the percentage of trees that clustered together based on the bootstrap test (1000 replicates) are shown next to the branch nodes.
The other well-supported group included E. coli sequences (eight nuclei per cyst). Within this group, two clades were distinguished: B and C. Clade B included subtype II E. coli sequences and clade C included subtype I. Our newly obtained sequences of E. coli were part of the subtype I group.
The sequence obtained for E. histolytica (ON712729) could not be aligned because it had a lower coverage.
DiscussionThe molecular epidemiological information on Entamoeba species is still scarce in Argentina. This study provides new genetic data from E. coli, E.dispar and E. histolytica. The phylogenetic relationship inferred from SSU rRNA gene data was consistent with the classification of Entamoeba species based on the number of nuclei observed in the mature cysts.
In this study, the overall Entamoeba prevalence determined by morphological and molecular methods was 12.4%, being more prevalent in Buenos Aires than in Misiones (14.8% vs. 10.6%). In previous studies performed in Buenos Aires, the prevalence of Entamoeba spp. ranged from 8.9 (7/79) to 33.7% (27/80) and E. coli was the most prevalent species. In these studies, the diagnosis was made by conventional methods, such as formol-ethyl concentration and flotation methods, and detected E. coli as the most prevalent species (Supplementary Table 1). On the other hand, only one study detected E. histolytica in Buenos Aires by microscopic-based techniques (Ritchie's sedimentation, Willis's flotation, and trichrome staining)9. However, formalin-fixed samples were analyzed in this study, which means that the morphology of the trophozoites was not preserved. Therefore, it cannot be stated that the authors had correctly identified this protozoan.
In Misiones, the prevalence of Entamoeba spp. ranged from 70.6 (153/218) to 7.4% (36/483) and E. coli was also the most prevalent species (Supplementary Table 1). In addition, different studies revealed the presence of E. histolytica, E. dispar and E. hartmanni in native populations by molecular characterization of the SSU rRNA gene4,43.
As described above, previous studies have reported a wide range of prevalence of Entamoeba infection in both provinces. Since this protozoan is primarily transmitted through the fecal–oral route21, the prevalence of the infection may be more closely related to socioeconomic factors and individual habits rather than to the edaphoclimatic characteristics of the provinces analyzed4,25. In this study, the prevalence was slightly higher in Buenos Aires than in Misiones. While individuals in both provinces lived in impoverished conditions, the residents of Buenos Aires experienced a lack of access to piped water, while the majority of those living in Misiones relied on well water for consumption. In this sense, the source of drinking water has been identified as a significant risk factor for Entamoeba infections3. Therefore, these characteristics, rather than the environmental factors, may have strongly influenced the prevalence of this parasite infection in these areas.
Differential diagnosis of the pathogenic E. histolytica and nonpathogenic Entamoeba species is crucial for treatment decisions and public health knowledge15. In addition, the accurate diagnosis of non-pathogenic amebas is important since the infection with these protozoans indicates fecal exposure (e.g. by food or drinking water), and may suggest possible exposure to pathogenic organisms11. In developing countries of the Americas, amebiasis is typically diagnosed by identifying parasite cysts or motile trophozoites by microscopic examination of the suspected fecal samples10,31. The disadvantages of this conventional method include its low sensitivity and specificity, with false positive results due to the presence of other Entamoeba species10. As a consequence, several other diagnostic procedures, based on parasite culture, serologic tests, antigen detection, and PCR, have been developed17. In this sense, PCR has been approved by the World Health Organization as the method of choice for the diagnosis of E. histolytica16.
In this work, a combination of microscopic analyses and PCR assays, with high sensitivity and specificity to detect Entamoeba species, developed by Gonin and Trudel15 and Verweij et al.39 were performed. Thus, E. coli subtype I was detected in both Buenos Aires and Misiones populations, and a case of E. histolytica was detected in Buenos Aires. Regarding E. coli, it is a species complex characterized by the extensive diversity of cryptic sequences in the genes of the small subunit of ribosomal RNA, which has led to the recognition of two distinct subtypes (ST1 and ST2)36. There are scarce molecular data on this species in South America. In fact, this is the first study to molecularly characterize isolates of this protozoan species from Argentina. E. coli ST1 and ST2 were found in Brazil, while ST2 was recorded in Perú and Ecuador36. Stensvold et al.37 have shown that ST2 was more common in samples collected outside Europe, whereas ST1 showed no geographical restriction. Similarly, this study provides the first molecular characterization of an E. histolytica isolate from Buenos Aires, which was obtained from a 24-year-old woman born in Paraguay but residing in Buenos Aires since 2015. Although the patient was asymptomatic, she received medical treatment and repeat parasitological testing was performed to confirm that she was parasite-free.
The same diagnostic approach was performed to diagnose Entamoeba species in a native community from Misiones43. This methodology permitted the establishment of a more reliable diagnosis, detecting infections with E. coli, E. dispar and E. hartmanni43.
The importance of an accurate diagnosis resides not only in treating the patients properly, but also in better understanding the epidemiology of amebiasis, which is a leading cause of severe diarrhea worldwide and accounts for 2.2 million disability-adjusted life years and around 55500 annual deaths38. Previous studies have shown that factors, such as the consumption of untreated contaminated water, sewage system, inadequate hygienic practices and social determinants were associated with a high prevalence of E.histolytica infections3,25. As mentioned earlier, in this study, most of the individuals analyzed were characterized as living in poor socioeconomic conditions, without access to drinking water and a sewage network. In this sense, even though the prevalence of E. histolytica was low (0.2%), the overall prevalence of Entamoeba spp. infections (12.4%) may indicate the fecal contamination of water or food that people consumed.
ConclusionsSince the microscopic analyses allowed to screen all intestinal parasite infections and PCR was found to be a more reliable source of E. histolytica detection, a combination of these techniques was performed to study the prevalence of Entamoeba species in populations from Argentina. Among the Entamoeba complex species, E. dispar was the most prevalent. The first molecular record of E. histolytica and characterization of isolates of E. coli and E. dispar were supplied. Moreover, this study increases the information on the distribution of these species in Argentina and the region of the Americas.
Author's contributionsAS conceived the study. AS participated in study design and molecular analyses, and analyzed and interpreted the data. AS performed the sampling, and parasitic examinations and wrote the draft of the manuscript. MLZ performed the sampling and parasitic examinations and contributed to discussing the results. GTN helped with parasitic examinations, coordinated the study and contributed to discussing the results. All authors read, revised and approved the final manuscript.
FundingThis work was supported by the Agencia Nacional de Promoción Científica y Tecnológica [PICT-2018-03763] and the Universidad Nacional de La Plata [UNLP 11/N881 and UNLP 11/N942].
Conflict of interestNone declared.
We are grateful to Richard Bradbury (CDC, USA) for providing Entamoeba histolytica and E. dispar DNA samples. We would like to thank the families who participated in this study. We are also grateful to Nicolas Viera for his technical assistance and Lucas Garbin for revising and proofreading the manuscript.