Endophytic fungi are ubiquitous and live within host plants without causing any noticeable symptoms of disease. Little is known about the diversity and function of fungal endophytes in plants, particularly in economically important species. The aim of this study was to determine the identity and diversity of endophytic fungi in leaves, stems and roots of soybean and corn plants and to determine their infection frequencies. Plants were collected in six areas of the provinces of Buenos Aires and Entre Ríos (Argentina) two areas were selected for sampling corn and four for soybean. Leaf, stem and root samples were surface-sterilized, cut into 1cm2 pieces using a sterile scalpel and aseptically transferred to plates containing potato dextrose agar plus antibiotics. The species were identified using both morphological and molecular data. Fungal endophyte colonization in soybean plants was influenced by tissue type and varieties whereas in corn plants only by tissue type. A greater number of endophytes were isolated from stem tissues than from leaves and root tissues in both species of plants. The most frequently isolated species in all soybean cultivars was Fusarium graminearum and the least isolated one was Scopulariopsis brevicaulis. Furthermore, the most frequently isolated species in corn plants was Aspergillus terreus whereas the least isolated one was Aspergillus flavus. These results could be relevant in the search for endophytic fungi isolates that could be of interest in the control of agricultural pests.
Los hongos endófitos son ubicuos y se encuentran en el interior de los tejidos de las plantas de manera asintomática. Se sabe muy poco acerca de la diversidad y la función de estos hongos, particularmente en especies de importancia económica. El objetivo de este trabajo fue determinar la diversidad y la frecuencia de colonización de hongos endófitos en raíces, tallos y hojas de 2 variedades de maíz y de 4 variedades de soja; las muestras se tomaron de 6 áreas diferentes ubicadas en las provincias de Buenos Aires y Entre Ríos (Argentina). Con un bisturí estéril se obtuvieron porciones de 1 cm2 de raíz, tallo y hoja, que fueron colocados en placas con agar papa dextrosa más antibiótico.
Las especies de hongos fueron identificadas a partir de características morfológicas y moleculares. La colonización de hongos endófitos en soja estuvo influenciada por la variedad y por el tipo de tejido, en tanto que en el maíz solo hubo influencia del tipo de tejido. El mayor número de endófitos se encontró en los tallos de ambas especies. El aislamiento más frecuente en todas las variedades de soja fue Fusarium graminearum y el menos frecuente Scopulariopsis brevicaulis. En ambas variedades de maíz la especie con mayor frecuencia de aislamiento fue Aspergillus terreus y la de menor fue Aspergillus flavus. Estos resultados son relevantes para la búsqueda de especies de hongos endófitos que podrían ser de interés en el control de plagas agrícolas.
By the year 2020, the supply of food especially of cereals, will have to increase about 70% in developing countries to secure food for the projected population of 6500 million people. It is expected that most of this increase in food supply will come from developing countries33. Soybean and corn are the extensive major crops in Argentina, providing a high percentage of the basic food needs of the population. The most important crops in Argentina are soybean and corn with 18 and 3.4million sown hectares, respectively19.
Symptomless internal colonization of healthy plant tissues by fungi is a widespread and well-documented phenomenon. Increasing interest in the ecological roles of these fungi has stimulated research in recent years since they might have plant growth – promoting activity13. Endophyte is an all-encompassing topographical term that includes all those organisms that during a variable period of their life symptomlessly colonize the living internal tissues of their hosts24. It is hypothesized that fungal endophytes, in contrast to known pathogens, generally have far greater phenotypic plasticity and thus more options to interact with their host than pathogens27. Since the 1970s several reports have shown that these fungal endophytes play important roles in protecting their host against predators and pathogens25. Endophytic fungi that infect plants are ubiquitous in all environments studied7,24,28. Although the diversity and function of fungal endophytes that infect grasses are well documented, little is known about the diversity and function of fungal endophytes in plants, particularly in economically important species13,26. Some fungal endophytes can reportedly reduce plant diseases and enhance plant growth and may be the basis for emerging methods to improve plant growth and production12,17,18,20. For example treatment of soybean [Glycine max L. (Merr)] with culture filtrate from the endophyte Cladosporium sphaerospermun increased plant height3,11,20,21. Although soybean and corn are major world crops, there is very limited knowledge of their fungal endophyte community.
The goal of this study was to isolate fungal endophytes from leaves, stems and roots of four soybean and two corn varieties grown in agricultural sites of the provinces of Buenos Aires and Entre Rios, Argentina, and to determine their colonization frequencies.
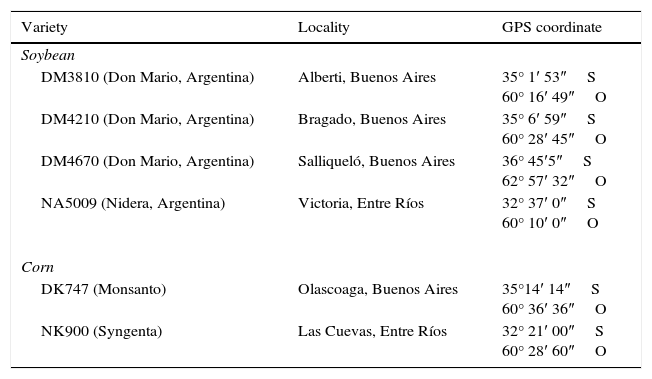
Materials and methodsSample collectionThe plants were collected during January and February 2013 in six locations in the soybean and corn cropping area of the provinces of Buenos Aires and Entre Rios (Argentina)5 (Table 1). The region's climate is temperate with an average temperature of 17°C and an average annual rainfall of 1000mm. Two areas were selected for sampling corn and four for soybean sampling, since soybean varieties are more predominant in Argentina. Plants of both species were grown in monoculture fields with a history of annual corn and soybean rotation. Ten plants without symptoms of disease were randomly selected from each plot in soybean cultivars DM 3810, DM 4210, DM 4670 (Don Mario Co., Buenos Aires province, Argentina), NA 5009 (Nidera Semillas Co., Buenos Aires province, Argentina) and ten plants were selected from corn cultivars NK 900 (Sygenta Semillas Co., Argentina) and DK 747 (Dekald®, Argentina). All samples were collected at 60–70 days after germination, cut at the soil line, immediately placed on ice and stored up for 72h at 4°C until processed according to Impulliti and Malvick13.
Locations where different soybean and corn plants were sampled
| Variety | Locality | GPS coordinate |
|---|---|---|
| Soybean | ||
| DM3810 (Don Mario, Argentina) | Alberti, Buenos Aires | 35° 1′ 53″S 60° 16′ 49″O |
| DM4210 (Don Mario, Argentina) | Bragado, Buenos Aires | 35° 6′ 59″S 60° 28′ 45″O |
| DM4670 (Don Mario, Argentina) | Salliqueló, Buenos Aires | 36° 45′5″S 62° 57′ 32″O |
| NA5009 (Nidera, Argentina) | Victoria, Entre Ríos | 32° 37′ 0″S 60° 10′ 0″O |
| Corn | ||
| DK747 (Monsanto) | Olascoaga, Buenos Aires | 35°14′ 14″S 60° 36′ 36″O |
| NK900 (Syngenta) | Las Cuevas, Entre Ríos | 32° 21′ 00″S 60° 28′ 60″O |
Endophytic fungi were isolated according to the protocols described by Pimentel et al.25. All leaf, stem and root samples were washed twice in distilled water, then surface-sterilized by immersion for 1min in 70% (v/v) ethanol, 4min in sodium hypochlorite (3%, v/v available chlorine) and then washed three times in sterilized distilled water for each time. After surface sterilization, samples were cut into 1cm2 pieces with a sterile scalpel and aseptically transferred to plates containing potato dextrose agar (PDA, Britania S.A., Buenos Aires, Argentina) to which a 0.1% stock consisting of 0.02g of each of two antibiotics (chloramphenicol and streptomycin) dissolved in 10ml sterile distilled water was added, followed by filter sterilization through a 0.2-μm filter (Syringe filter sterile, E-Chrom Tech, Taiwan); 1ml of this was added to each litre of medium, to suppress bacterial growth31. Aliquots from the third wash were plated onto PDA to check that surface sterilization had been effective. A total of 1080 fragments were plated (18 from each of the 60 plants investigated). To facilitate isolation of endophytic fungi, the plates were incubated in the dark at 25°C. The plates were checked everyday for up to ten days after incubation and any fungi present was isolated, purified and then maintained at 4°C on PDA slopes for further identification. Percentage colonization was defined for each variety as the total number of fragments colonized by fungi in relation to the total number of fragments×10025.
The species were identified using both morphological and molecular data.
Morphological identification of the isolates was done by growing them on PDA plates or in microculture14 and examining the colonies for asexual or sexual reproductive structures using optical microscopy and taxonomic keys. Species were identified according to Leslie and Summerell15 and Domsch et al.8.
Genomic DNA of monosporic cultures was obtained according to Stenglein and Balatti29. To confirm morphological identifications, a PCR was carried out in an XP thermal cycler (Bioer Technology Co, Hangzhou, China) to amplify the ITS rDNA region using primer pairs ITS5 (5′-GGAAGTAAAAGTCGTAACAAG G-3′) and ITS4 (5′-TCCTCC GCT TATTGATATGC-3′)32. For Fusarium species confirmation the translation elongation factor (EF-1α) region was amplified using primers EF1 (5′-ATGGGTAAGGA(A/G)GACAAGAC-3′) and EF2 (5′-GGA(A/G)GTACCAGT(G/C)ATCATGTT-3′)22. The PCR reactions, the fragment purifications and sequencing were performed according to Canel et al.6 and Stenglein et al.30.
The similarities of the fragment with previously published sequence data were examined with BLASTn1 on the NCBI web page.
Diversity was assessed using the Shannon Index16 (for the cultivars and tissue types).
The differences between fungi isolates and frequency of colonization for the different varieties were tested using two-way analysis of variance (ANOVA) and their means were compared by the LSD test (p<0.05) using the Infostat software.
ResultsFrom the soybean plants sampled, 11 fungal species were isolated and identified using both morphological and molecular data. In all soybean cultivars, Fusarium graminearum was the most frequently isolated species sampled while Scopulariopsis brevicaulis was the least frequently isolated one (Table 2). Furthermore, in the corn plants sampled, 7 fungal species were isolated (six species belonging to Ascomycota and one to Zygomycota (Table 3)), being Aspergillus terreus the most frequently isolated species and Aspergillus flavus the least frequently isolated one (Table 3). All endophytic fungal species were deposited in the strain culture collection of the Spegazzini Institute, La Plata, Argentina (LPSC) (Tables 2 and 3).
Colonization percentage of different fungal species isolated from roots, stems and leaves from four different soybean cultivars sampled
| Fungal species | Colonization percentage (%) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DM3810 | DM4210 | DM4670 | NA5009 | |||||||||
| Root | Stem | Leave | Root | Stem | Leave | Root | Stem | Leave | Root | Stem | Leave | |
| LPSC1187 Alternaria alternata (Fr.) Keissl | – | – | – | – | 25 | 26.6 | – | – | – | – | – | – |
| LPSC1186 Arthrinium phaeospermun (Corda) Ellis | – | – | – | – | 20 | 33.3 | – | – | – | – | – | – |
| LPSC1181 Aspergillus niger Tiegh | – | 10 | 38.3 | – | 13.3 | 41.6 | – | – | – | – | – | – |
| LPSC1182 Clonostachys rosea (Link) Schroers | 16 | – | – | – | – | – | – | – | – | – | 15 | 30 |
| LPSC1178 Curvularia lunata (Wakker) Boedijn | – | 6.6 | 46.6 | – | 38.3 | – | – | – | – | – | 38.3 | – |
| LPSC1188 Fusarium graminearum Schwabe | 20 | 45 | – | 3.3 | 10 | 30 | 20 | 20 | 10 | 11.6 | 25 | 31.6 |
| LPSC1184 Fusarium equiseti (Corda) Sacc | – | – | 58.3 | – | – | – | – | – | – | – | – | – |
| LPSC1191 Fusarium oxysporum Schlecht | – | 10 | 21.6 | – | – | – | – | 33.3 | 8.3 | – | – | – |
| LPSC1185 Macrophomina phaseolina (Tassi) Goid | 13.3 | 28.3 | – | – | – | – | 10 | 13.3 | 28.3 | – | – | – |
| LPSC1189 Scopulariopsis brevicaulis(Sacc) Bainier | – | – | – | – | – | – | – | 41.6 | – | – | – | – |
| LPSC1179 Trichoderma saturnisporum Hammill | – | – | – | – | – | – | – | – | – | 10 | 16.6 | 38.3 |
Colonization percentage of different fungal species isolated from roots, stems and leaves from two different corn cultivars sample
| Fungal species | Colonization percentage (%) | |||||
|---|---|---|---|---|---|---|
| DK747 | NK900 | |||||
| Root | Stem | Leaf | Root | Stem | Leaf | |
| LPSC1183 Aspergillus flavus Link | 8.3 | 35 | – | – | – | – |
| LPSC1180 Aspergillus terreus Thom | 8.3 | 30 | 28.8 | 10 | 43.3 | – |
| LPSC1193 Bipolaris tetramera (McKinney) Shoemaker | – | 31.6 | – | 21.6 | 23.3 | – |
| LPSC1223 Clonostachys rosea (Link) Schroers | – | – | – | – | 18.3 | 31.6 |
| LPSC1224 Fusarium graminearum Schwabe | – | – | – | – | 28.3 | 18.3 |
| LPSC1190 Fusarium proliferatum (Matsush) Nirenberg | 26.6 | 43.3 | 28.3 | – | 25 | 26.6 |
| LPSC1192 Mucor circinelloides Tiegh | – | 25 | – | 10 | 33.3 | – |
With regard to fungal diversity in soybean plants, cultivar DM3810 showed the highest diversity while in cultivar NA5009 we observed the lowest diversity, with a Shannon's index of 3.09 and 1.93, respectively. Moreover, corn cultivar DK747 showed the highest fungal diversity whereas the lowest diversity was observed in cultivar NK900, with a Shannon's index of 1.84 and 1.67, respectively. Furthermore, based on the Shannon's index, the greatest fungal diversity in all the soybean cultivars sampled was observed in the stems and then in the leaves whereas the lowest diversity occurred in the roots, except for cultivar DM4670, where we observed increased diversity in the roots rather than in the leaves with a Shannon's diversity index of 1.35 and 1.18, respectively. With respect to the fungal diversity in the roots, stems and leaves of corn plants, most fungal diversity was observed in the stems of both DK747 and NK900 cultivars with a Shannon's diversity index of 2.27 and 2.49, respectively, then in leaves and the lowest diversity occurred in roots.
Fungal isolates identification was confirmed at molecular level and submitted to GenBank (Accession numbers: KF753941–KF753956). Fungal endophyte colonization in soybean plants was influenced by the cultivars, showing significant differences between varieties (F=4.17, df=3, p=0.0063), fungi isolates (F=6.93, df=10, p<0.0001) and in the interaction among them (F=7.12, df=30, p<0.0001) (Fig. 1). Corn plants showed no significant differences between cultivars (F=1.34, df=1, p=0.2500), however, they did instead among fungi isolates (F=4.07, df=6, p=0.0009) and in the interaction among cultivars and fungi isolates (F=4.12, df=6, p=0.0008) (Fig. 2).
Studies of fungal endophytes in many environments are an active area for research; however, the endophytes in soybean and corn have never been systematically characterized. This work expands our understanding of endophytic fungi in soybean and corn plants. We have focused on roots, stems and leaves because many soybean and corn pathogens commonly colonize these organs. The fungal endophytes identified in this study are not known to be soybean and corn pathogens, and the functional associations between these fungi and soybean and corn plants are unknown. Furthermore, it is important to mention that none of the plants used in this study had symptoms of disease. The most prevalent endophytic fungal species isolated in the organs (root, stem and leaf) in all soybean cultivars was F. graminearum and in the two corn cultivars was A. terreus. Pimentel et al.25; Impullitti and Malvick13 found that Cladosporium was the endophytic fungal genus most frequently identified from leaves and stems of soybeans grown in Brazil and Minnesota, USA whereas Pan et al.23 found this genus in leaves and stems from corn in Minnesota, USA. In this study only one species of Zygomycota was isolated in maize plants; species from this Phylum were isolated as endophytes in Dactylis glomerata L. and other plants18.
The endophytic fungal species detected in plants may be influenced by many factors, including the type of tissue sampled, the time when plants were assayed, perhaps the climate and location in which they were grown13, whether the plant is grown in a monoculture or polyculture, the plant age or cropping history of the field2,4,9,27. Soybean and maize used in this study were grown in a monoculture and in fields that had a history of corn, wheat and soybean rotations.
Fisher et al.10 observed that parts of corn-stems nearer to the soil showed a lower incidence of fungal infection and explained that this probably was due to these parts of the stem having an increased frequency of bacteria that inhibited fungal colonization. This could explain why we obtained the greatest number and diversity of isolates from stems in different soybean and corn cultivars, than in leaves and roots.
A greater number of fungi such as endophytes in stems were also observed in soybean in Brazil whereas endophytic bacteria in maize were found in the USA10,25. These studies also suggested that endophytes may exclusively colonize certain tissues, for example, Colletotrichum was only isolated from soybean leaves and not from the stems cultivated in Brazil25. In our study, Fusarium equiseti was only isolated from leaves of soybean plants and S. brevicaulis only from stems.
Endophytes may be important organisms to improve a sustainable production of crops, although their identities and functions in a range of plants are just beginning to be revealed. This is the first time that we study the natural endophytes placed in roots, stems and leaves of the main soybean and maize cultivars in Argentina. Species could be determined by classical taxonomy and the use of molecular techniques for each of the isolates obtained. In addition, we determined the colonization percentage, the fungal diversity in the different organs of every plant studied, the differences between fungi isolates and the frequency of colonization for different varieties using the ANOVA analysis. Future research should be conducted to determine which of these fungal natural endophytes could be used for both biological control and plant growth promotion.
Ethical disclosuresProtection of human and animal subjectsThe authors declare that no experiments were performed on humans or animals for this investigation.
Confidentiality of dataThe authors declare that no patient data appears in this article.
Right to privacy and informed consentThe authors declare that no patient data appears in this article.
Conflict of interestThe authors declare that they have no conflicts of interest.
This study was partially supported by Consejo Nacional de Investigaciones Científicas y Tecnológicas (CONICET PIP0009), Agencia de Promoción Científica y Tecnológica (PICT 2013-0543), Comisión de Investigaciones Científicas de la Provincia de Buenos Aires (CICPBA), Universidad Nacional de La Plata (UNLP, 11/N 773) and Rizobacter Argentina S.A.