The objectives of this study were: (1) to estimate STEC frequency in hide and carcass samples taken from beef slaughterhouses supplying the domestic market in Argentina, (2) to establish the pheno-genotypic characteristics of STEC and non-toxigenic Escherichia coli of serogroups O26, O45, O103, O121, O111, O145 or O157 isolated from the analyzed samples and, (3) to study their clonal relatedness. Sixty hides and 60 carcasses were analyzed. At the screening step, 48% of hide and 80% of carcass samples tested positive for the stx gene by endpoint PCR. The STEC isolation rate was 5% for hides and 8% for carcasses. The isolation rate of STEC-positive for O26, O45, O103, O111, O145 or O157 serogroups was 0% for hides and 2% for carcasses. With the purpose of studying the clonal relatedness of isolates, macrorestriction fragment analysis by pulsed-field gel electrophoresis was performed. The results indicated cross-contamination between hides and between carcasses of animals in the same lot and, that the origin of carcass contamination was their own hide, or the hides of other animals in the same lot. The high detection rate at the screening step, especially in carcasses, and the evidence of cross-contamination show the need to apply additional in-plant intervention strategies aimed at preventing carcass contamination.
Los objetivos del presente estudio fueron tres: 1) estimar la frecuencia de Escherichia coli productor de toxina Shiga (STEC) en muestras de cuero y carcasa de bovinos en frigoríficos de consumo interno de Argentina; 2) realizar la caracterización feno-genotípica de las cepas STEC y de Escherichia coli no toxigénicas pertenecientes a los serogrupos O26, O45, O103, O121, O145 u O157 aisladas a partir de las muestras analizadas; 3) establecer la relación clonal de ese conjunto de cepas. Se analizaron 60 cueros y 60 carcasas. En la etapa de tamizaje, el gen stx se detectó en el 48% de las muestras de cuero y en el 80% de las muestras de carcasa por una PCR de punto final. La frecuencia de recuperación de cepas STEC fue del 5% en cueros y del 8% en carcasas, y la de cepas STEC positivas para los serogrupos O26, O45, O103, O121, O111, O145 u O157 fue del 0% en los cueros y del 2% en las carcasas. La relación clonal de las cepas aisladas se investigó a través de electroforesis de campo pulsado y análisis de los patrones de macrorrestricción generados. Los resultados demostraron la existencia de contaminación cruzada entre cueros y carcasas de animales pertenecientes a un mismo lote, y también que el origen de la contaminación fue el propio cuero del animal o el cuero de otros animales pertenecientes al mismo lote. Los altos porcentajes de detección en la etapa de tamizaje, especialmente en carcasas, y la evidencia de contaminación cruzada ponen de manifiesto la necesidad de evaluar la implementación de estrategias de intervención tendientes a evitar la contaminación de carcasas.
Shiga toxin-producing Escherichia coli (STEC) can cause bloody diarrhea, hemorrhagic colitis and hemolytic uremic syndrome (HUS). In Argentina, HUS is endemic, with an incidence of approximately 400 cases per year17.
STEC includes all E. coli carrying the stx gene. Some STEC harbor a pathogenicity island inserted in the genome known as locus of enterocyte effacement (LEE), which contains the eae gene encoding intimin and responsible for attaching and effacing lesions in the gut mucosa. E. coli O157:H7 is the most common serotype associated with human diseases worldwide, including Argentina. However, the recent increases in the number of outbreaks and sporadic cases were due to non-O157 STEC serogroups O26, O45, O103, O111, O121 and O14510. Their low infectious dose and their association with the consumption of raw or undercooked beef products, led the United States Department of Agriculture – Food Safety and Inspection Service (USDA-FSIS) (2014) to declare these six serogroups, known as the “Big Six”, as adulterants in raw, non-intact beef products and product compounds since 201230. In Argentina, SENASA (National Animal and Plant Health Service and Food Quality) is the federal agency in charge of the sanitary oversight of meat, meat products and slaughterhouses. By issuing notice 4032/12, SENASA, in fact, adopted the recommendations of USDA-FSIS included in the Microbiology Laboratory Guide 5B.0528.
The origins and subsequent rate at which carcass contamination occurs have not been well established. Many studies support the hypothesis that the hide is the major source of carcass contamination by E. coli O157:H7 and non-O157 STEC2,13. Elder et al.7 suggested that cross contamination may also occur when large numbers of infected animals are being processed. Contamination could occur through direct contact with personnel, knives or other equipment7.
For the purpose of reducing STEC on carcass the US beef industry applies antimicrobial interventions at different points of the process line32. In the Argentinean industry, lactic acid was recently approved for use and remains the only chemical that may be applied to carcass as antimicrobial intervention28. However, to the best of our knowledge, it is not currently used at slaughterhouses that supply the domestic market of Argentina.
The aims of this study were: (1) to estimate STEC frequency in hide and carcass samples taken from beef slaughterhouses that supply the domestic market in Argentina, (2) to establish the pheno-genotypic characteristics of STEC and non-toxigenic E. coli of serogroups O26, O45, O103, O121, O111, O145 or O157 strains isolated from the analyzed samples and, (3) to study their clonal relatedness.
Materials and methodsSample collectionTwo slaughterhouses were visited five times from November 2014 to September 2015 (total number of visits: 10). On each visit, 12 samples were taken, six from hides and six from carcasses. The hide and carcass samples were taken from the same animal and all animals belonged to the same lot.
Samples were collected using Speci-Sponges (Nasco, Fort Atkinson, WI) moistened with 25ml of buffered peptone water (BPW, Difco Laboratories). Hide samples were taken from a 1000cm2 area over the plate. Carcass samples consisted of an 8000cm2 area obtained by sampling two 4000-cm2 areas from one carcass side at the end of the process line31.
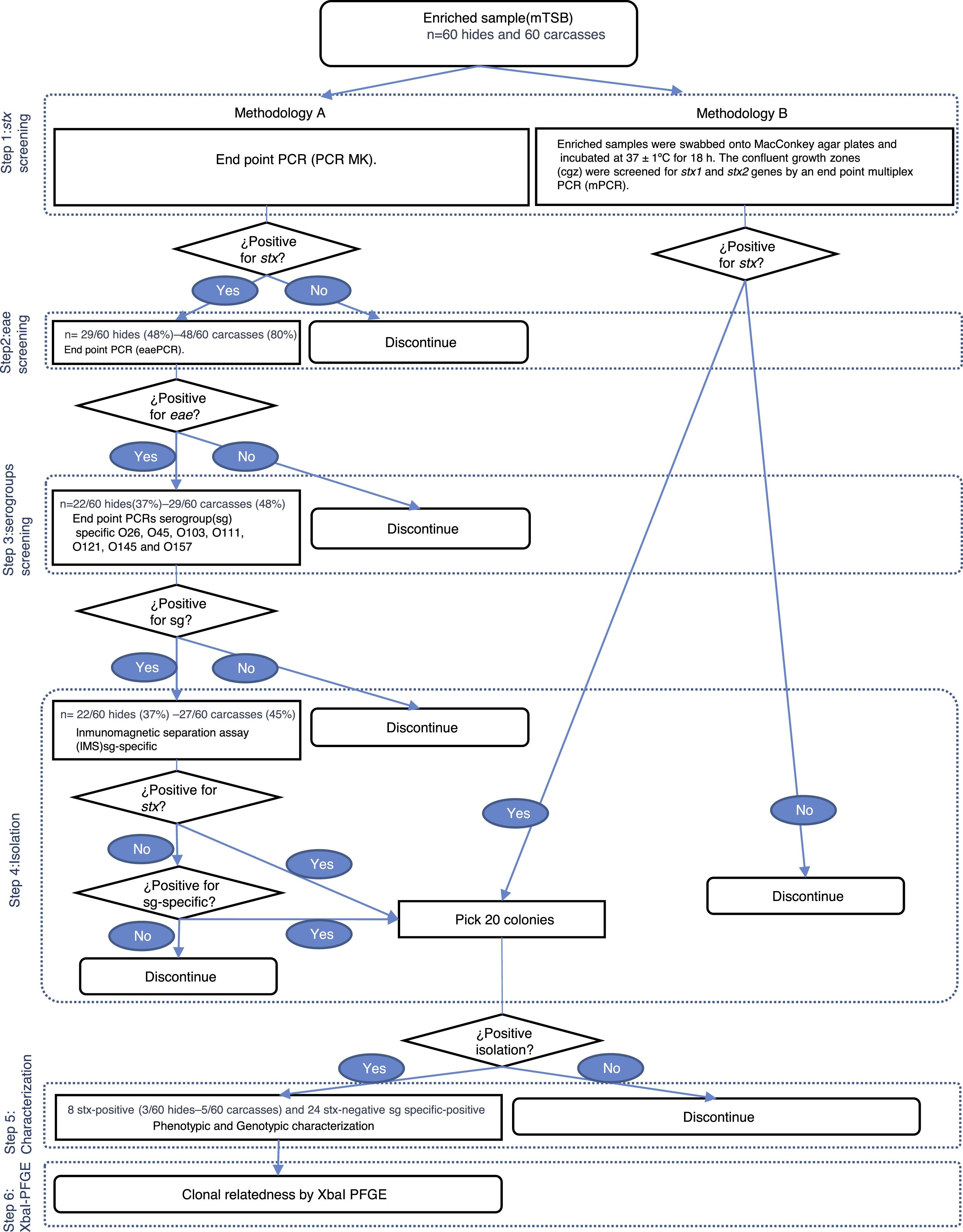
Detection and isolation of STECUpon arrival at the laboratory, 125ml of modified Triptone Soya Broth (mTSB; Oxoid, UK) were added to the Speci-Sponges and incubated at 42±1°C for 18h. Two methodologies, named A and B, were used. In Methodology (Met) A, enriched samples were analyzed for the presence of the stx gene by conventional PCR MK12. Positive samples for stx were tested for the presence of the eae gene by conventional PCR11. Positive samples for the stx and eae genes were tested for O26, O45, O103, O121, O111, O145 and O157 serogroups by 7 conventional PCRs4,6,9,14,21–23. If a positive signal was detected, samples were processed by serogroup-specific immunomagnetic separation assay (IMS; Dynal beads, Invitrogen, USA), according to the manufacturer's specifications. The separated cell/beads complexes were suspended in 100μl of phosphate buffered saline (PBS; BioRad, USA). Aliquots of 50μl of anti-E. coli O157 beads complexes were spread onto both Sorbitol MacConkey Agar (SMAC; Oxoid, UK) and ID-Chromagar (BioMérieux, France); aliquots of 50μl anti-E. coli O26, O45, O103, O111, O121 and O145 were spread onto both ChromAgar (CHOMagar, France) and SMAC using a sterile inoculation loop to obtain isolated colonies. The confluent growth zones were screened for the stx1, stx2 and serogroups genes (O26, O45, O103, O111, O121, O145 and O157) by endpoint PCRs. If the stx1, stx2 or serogroup genes were detected, 20 presumptive colonies were analyzed in order to isolate the bacteria harboring the genes of interest (Fig. 1).
In Met B, the enriched samples were swabbed onto MacConkey agar plates (MAC; Oxoid, UK) and incubated at 37±1°C for 18h. The confluent growth zones were screened for the stx1 and stx2 genes by endpoint multiplex PCR (mPCR)14. If the stx1 or stx2 genes were detected, 20 presumptive colonies were analyzed in order to isolate the bacteria that harbor the genes of interest (Fig. 1).
Phenotypic and genotypic characterization of isolatesConfirmation of isolates as E. coli was performed through biochemical test strip (BioMérieux API 20E®, France). Serotyping was conducted according to Orskov and Orskov19, using somatic (O1 to O182) antisera [Statens Serum Institute (Copenhague, Denmark)] and flagelar (H1 to H56) antisera [Denka-Seiken (Tokyo, Japan)]. The genotypic characterization included the detection of different virulent genes, such as eae, saa (STEC autoagglutinating adhesion)20, aggR25 (transcriptional activator of enteroaggregative E. coli) and ehxA (enterohemolysin)27 and the analysis of Stx1 and Stx2 variants according to Scheutz et al.26. Serotyping and the analysis of stx1 and stx2 variants were exclusively for the STEC strains, the genotypic characterization was conducted for all strains, STEC and non-toxigenic E. coli of serogroups O26, O45, O103, O121, O111, O145 or O157.
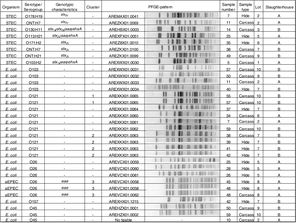
Clonal relatednessMacrorestriction fragment analysis by pulsed-field gel electrophoresis (PFGE) was performed using the Standard Operating Procedure for PulseNet PFGE of E. coli O157:H7 and non-O157 (CDC, 2013). Restriction digestion of DNA was carried out with the XbaI enzyme (Fermentas, EU). PFGE images of the gels were captured using a Doc-IT 2000 (Bio-Rad). The analysis of TIFF images was conducted using the BioNumerics version 5.1 software package (Applied Maths, Sint-Martens-Latem, Belgium). The Dice coefficient and the unweighted pair group method with arithmetic mean (UPGMA) were used to generate dendrograms with 1.5% tolerance values. The strains were grouped in a cluster when they showed identical XbaI PFGE pattern (100% similarity). The relationship shown among the strains by the BionNumerics analysis was visually confirmed.
ResultsDetection and isolation of STEC on hidesMet A showed that 48% (29/60) of the hide samples tested positive for stx whereas 37% (22/60) tested positive for the stx and eae genes. At the serogroup screening step all samples tested positive for, at least, one of the seven serogroups studied. The serogroups detected, in decreasing order of frequency, were: O121 (14/22; 63%), O103 (8/22; 36%), O26 (6/22; 27%), O145 (4/22; 19%), O157 (1/22; 5%), and O111 (0/22; 0%). Eleven E. coli strains were recovered by serogroup-specific IMS; however, none of them were toxigenic (Fig. 2). Met B showed that 13% (8/60) of the hide samples tested positive for stx whereas STEC strains were isolated in 5% (3/60) of the samples. As no STEC strain was isolated in Met A, the overall frequency of STEC in the hide samples was 5% (3/60) (Fig. 1).
Detection and isolation of STEC on carcassesMet A showed that 80% (48/60) of the carcass samples tested positive for stx whereas 48% (29/60) tested positive for stx and eae genes. At the serogroup screening step, 45% (27/60) tested positive for, at least, one of the seven serogroups studied. The serogroups detected, in decreasing order of frequency, were: O121 (23/27; 85%), O103 (8/27; 30%), O45 (4/27; 15%), O26 and O145 (1/27; 4%). Fourteen E. coli strains were recovered by serogroup-specific IMS; however, only 1 was toxigenic (Fig. 2). Met B showed that 32% (19/60) of the carcass samples were positive for stx and, STEC strains were isolated in 7% (4/60) of the samples. The overall frequency (Met A+B) of STEC in carcass samples was 8% (5/60) (Fig. 1).
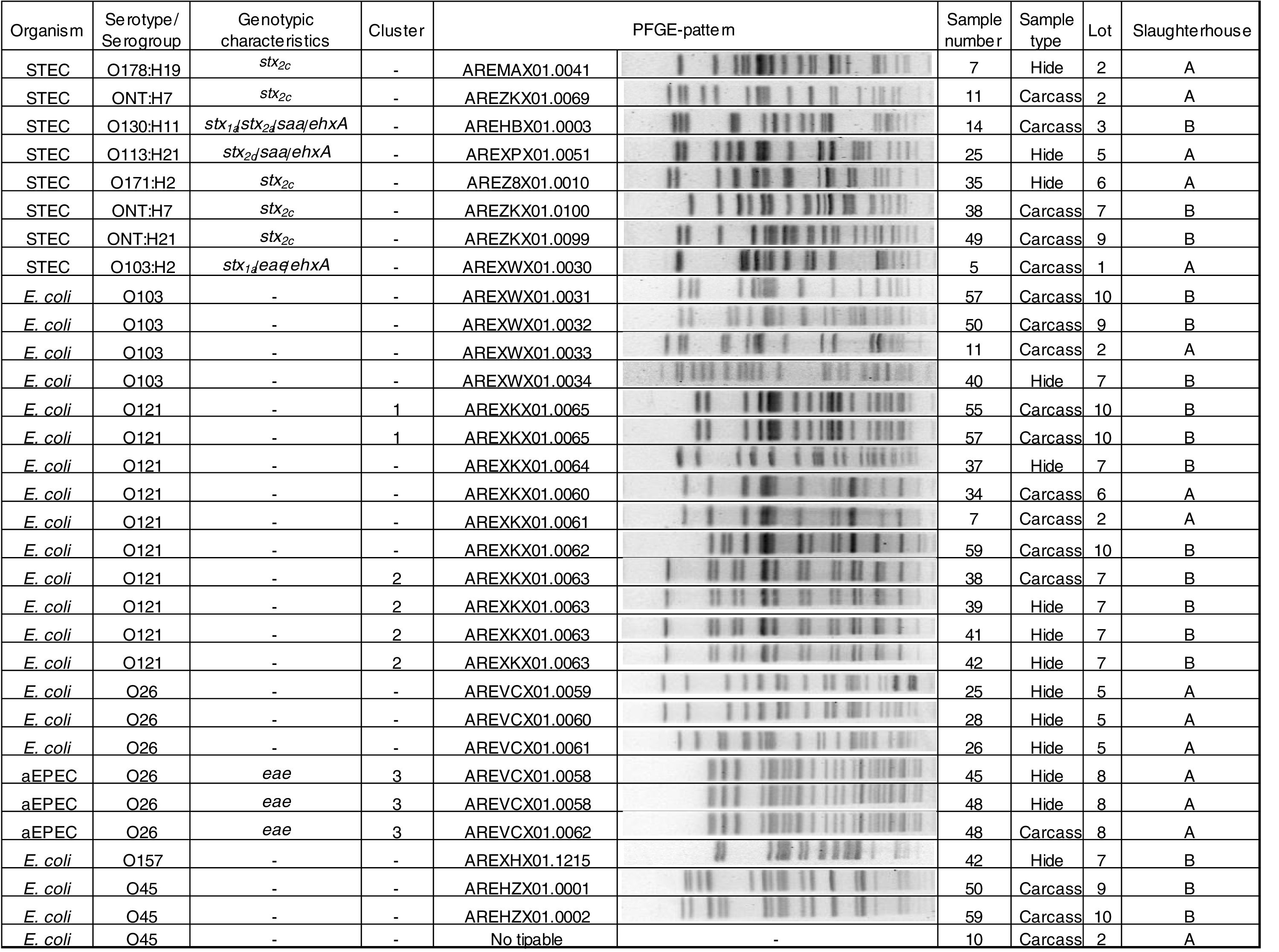
Phenotypic and genotypic characterizationThe 8 STEC strains isolated (3 from hides and 5 from carcasses) were characterized as: O103:H2 stx1a/eae/ehxA; O130:H11 stx1a/stx2a/saa/ehxA, O113:H21 stx2d/saa/ehxA, O178:H19 stx2c, O171:H2 stx2c, ONT:H21 stx2c and two strains ONT:H7 stx2c. Three O26 strains were aEPEC (atypical enteropathogenic E. coli). Twenty-one strains were negative for all the virulence factors studied and were the following: O121 (n=10), O103 (n=4), O26 (n=3), O45 (n=3), and O157 (n=1) (Fig. 2).
Clonal relatednessPFGE analysis showed 25 different patterns, including 3 clusters (strains with PFGE patterns with 100% similarity). The analysis of the patterns was performed by serogroup.
Cluster 1 included two E. coli O121 strains recovered from slaughterhouse B from carcasses of different animals in the same lot (lot 10). Cluster 2 included four E. coli O121 strains, also recovered from slaughterhouse B, from hides and carcasses of different animals in the same lot (lot 7). A 96% similarity was found between the patterns of two E. coli O121 strains recovered from slaughterhouse A from carcasses of different animals (sample number 34 and 7) and different lots (lots 2 and 6). Cluster 3 included three aEPEC O26 strains recovered from slaughterhouse A, from the hide and carcass of the same animal and from hides of two different animals in the same lot (lot 8) (Fig. 2).
DiscussionIn the present study, 48% of hides tested positive for stx using Met A at the screening step. Previous studies have reported rates as high as 97%5, 92%1 and 67%18. The isolation rate of STEC-positive serogroups included in the “Big Six” was be 0%, the same value reported by Monaghan et al.18. The isolation rate of STEC using Met B was 5%, similar to the 6% reported by Monaghan et al.18 but considerably lower than the 56% reported by Barkocy-Gallagher et al.1.
In carcasses, 80% tested positive for stx using Met A at the screening step; a value that was significantly higher than the stx-frequency detected in hides (48%). This may indicate that in-plant harvest practices and procedures were not successful at preventing cross contamination. Other studies reported frequencies of 97%1 and 33%5 in pre-evisceration carcasses. The isolation rate of the STEC-positive serogroup included in the “Big Six” was 2% (1/60), higher than the 0.4% reported by Monaghan et al.18 and the 0.1% reported by Masana et al.15. The isolation rate of STEC by Mets A and B was 8%; higher than the 1% reported by Monaghan et al.18 but, considerably lower than the 58% reported by Barkocy-Gallagher et al.1. The only highly pathogenic strain included in the “Big Six” was isolated using Met A. Met B showed higher STEC isolation rates; however, it was not useful for the recovery of highly pathogenic strains.
Several studies have been conducted in Argentina to estimate the occurence of non-O157 STEC on carcasses. Etcheverría et al.8 reported 12%, Masana et al.15 9%, Brusa et al.3 6% and Perez et al.24 0.4%. It is difficult to compare results among studies due to differences in animal types and ages, feeding regimen, sample size, detection and isolation procedures and, last but not less important, due to how efficient the manufacture practices were to prevent cross contamination at each slaughterhouse.
The prevalence of STEC O157 was 0% in both hides and carcasses. In Argentina, Masana et al.16 reported an isolation rate of 3% in carcasses. A possible explanation for this result might be the addition of novobiocin to the enrichment media and the fact that all samples were analyzed directly using IMS O157, skipping the screening step.
The high detection rates at the screening steps followed by very low isolation rates could be explained by the fact that the genes detected in each screening step could be harbored by different bacteria. The isolation of 24 strains positive for O26, O45, O103, O111, O121, O145 or O157 serogroup but negative for the stx gene is evidence of the afore mentioned. These results are consistent with Thomas et al.29 findings. Furthermore, highly pathogenic STEC is likely to be present at low concentrations, may be sublethally-injured and is usually accompanied by a large population of competent microflora, including other E. coli.
With regard to the PFGE pattern analysis, results indicated: (1) cross-contamination between hides and also between carcasses of animals in the same lot and, (2) that the origin of carcass contamination was its own hide, or the hides of other animals in the same lot. Same results were found by Thomas et al.29, Barkocy-Gallagher et al.2 and Brusa et al.3. Additionally, the high percentage of similarity (96%) between two E. coli recovered from carcasses during different visits may be due to persistent contamination at slaughterhouse A.
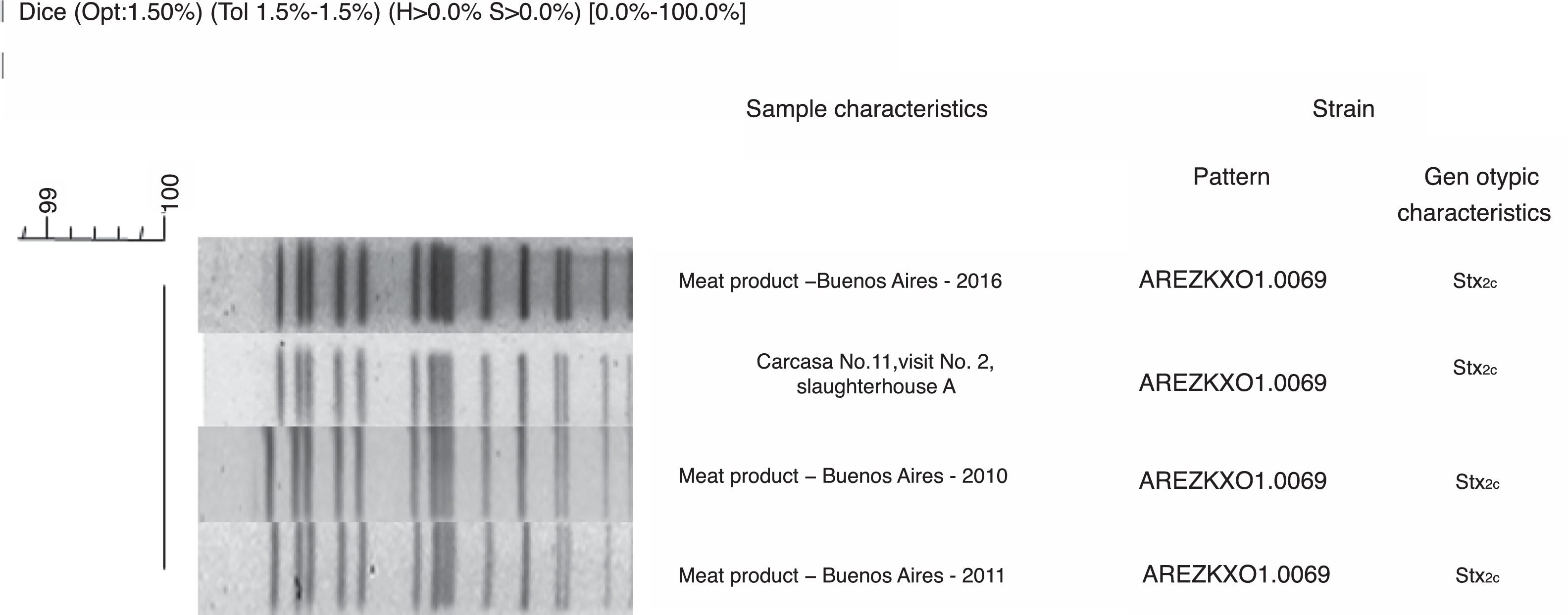
PFGE patterns were also compared with National Data Base of non-O157 STEC. A 95% similarity was found between STEC O103:H2 recovered from slaughterhouse A, from carcass sample number 5 in lot 1 and, the pattern of STEC O103:H2 recovered from a case of HUS in Buenos Aires, Argentina in 2010. This result could evidence that Argentinean cattle carry highly pathogenic STEC. However, since they are not related temporally or spatially, it could be described as a coincidence. A 100% similarity was found between STEC ONT:H7 recovered from slaughterhouse A, from carcass sample number 11 in lot 2 and the pattern of other three STEC ONT:H7 recovered from meat products in 2010, 2011, and 2016 in Buenos Aires City, Argentina (Fig. 3) which could mean that the strain had been circulating in Argentina for a long period of time.
The Xbal-PFGE analyses and the high frequency rates at the screening steps reported in this study, especially in carcasses, show the need for reviewing the procedures used in slaughterhouses supplying the domestic market in Argentina from the standpoint of their safety. It is important to promote the improvement at different safety levels and, also to evaluate the eventual implementation of antimicrobial intervention strategies.
These findings should be taken into account in order to improve the methodologies for the surveillance of foodborne pathogens that represent a risk to global health.
FundingThis project was supported by public funds of the Research Project ANAIyAI 1130042 of the Instituto Nacional de Tecnología Agropecuaria de Argentina (INTA) and the Instituto Nacional de Enfermedades Infecciosas “Dr. Carlos G. Malbrán” (INEI).
Conflict of interestNone of the authors of this paper has a financial or personal relationship with other people or organizations that could inappropriately influence or bias the content of the paper.
We are grateful to Geronimo Ortigoza for the technical assistance. We also thank the participating slaughterhouses for their contribution.