Metallo-β-lactamases (MBL) producing Pseudomonas aeruginosa isolates have been well characterized. Quinolones are commonly used in the treatment of carbapenem-resistant P. aeruginosa infections; however, data about PMQR in this species are scarce. The objective of this study was to report the simultaneous presence of qnrS and blaVIM-11 in P. aeruginosa, and to characterize the qnrS-harboring plasmid.
Thirty-eight carbapenem-resistant P. aeruginosa isolates were recovered from a hospital in Buenos Aires during 2012. Screening for MBL was assessed by the double disk synergy test using EDTA and carbapenem discs. Plasmid DNA extraction was performed by a method using phenol-chloroform. PCR followed by sequencing was carried out to determine each MBL and PMQR allele. PCR-BseGI-RFLP was performed to detect aac-(6′)-Ib-cr. The gyrA-QRDR was sequenced in those PMQR-harboring isolates. Plasmid incompatibility groups and addiction systems were characterized by PCR. The PMQR-carrying plasmid was sequenced using Illumina technology, annotated using RAST and manually curated.
Eleven/38 isolates were VIM producers (blaVIM-2 and blaVIM-11) while 1/38 harbored blaIMP-13. One isolate harbored blaVIM-11 and the PMQR qnrS1; however, both markers were located in different plasmids.
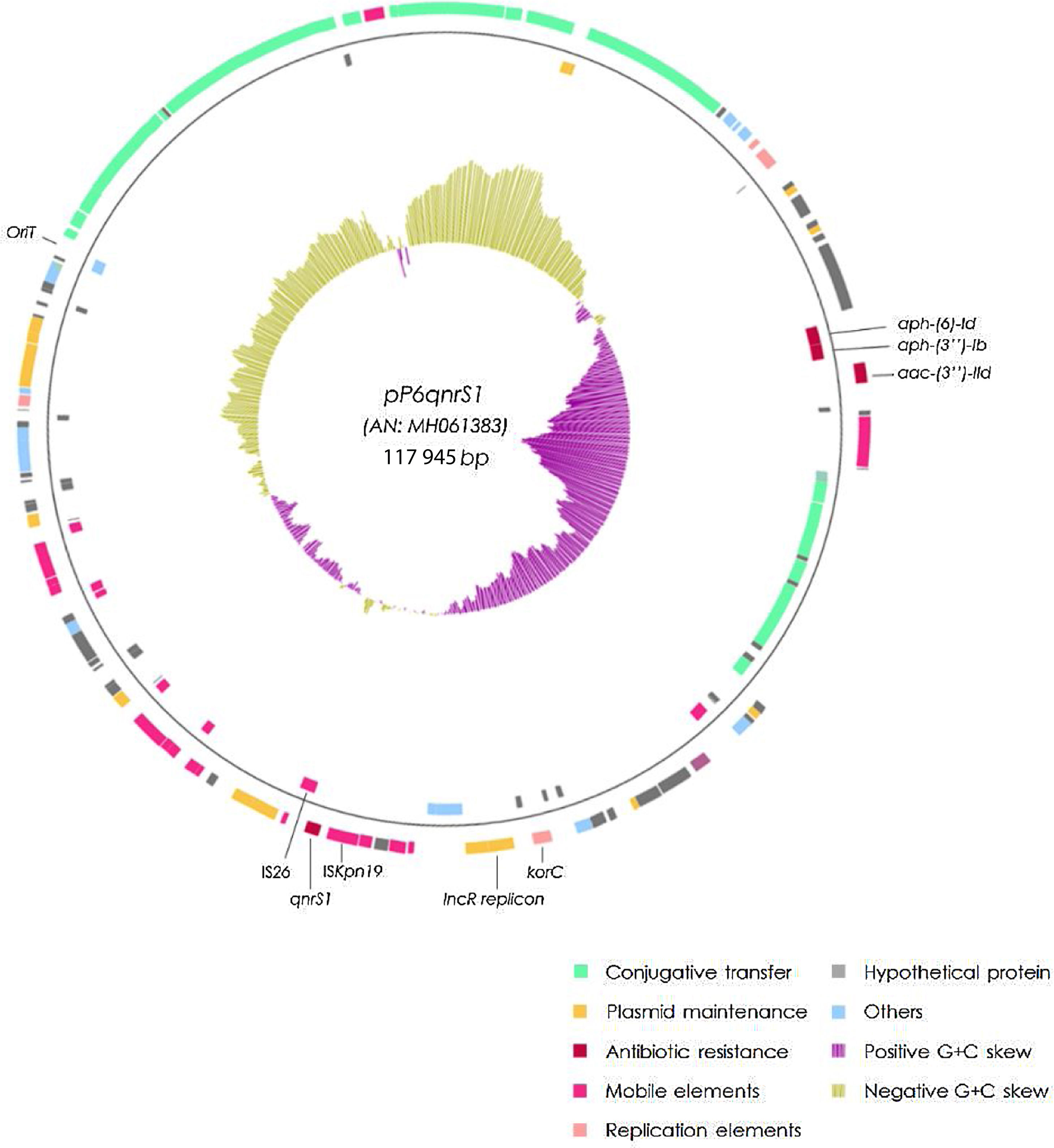
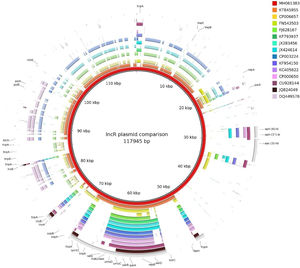
The qnrS1-harboring plasmid (pP6qnrS1) was 117945bp in size, presented 154 CDS and corresponded to the IncR group. In addition to qnrS1, it harbored several aminoglycoside resistance markers. Although pP6qnrS1 was non-conjugative, it presented an oriT which made it possible for this plasmid to be transferable.
This is the first report on P. aeruginosa carrying both blaVIM-11 and qnrS1, plus the first detection of an IncR plasmid in Argentina.
Las quinolonas son comúnmente utilizadas en el tratamiento de las infecciones producidas por Pseudomonas aeruginosa resistentes a carbapenems (PARC); aun así, la información sobre la resistencia a quinolonas mediada por plásmidos (PMQR) en esta especie es escasa. El objetivo de este trabajo fue reportar la presencia simultánea de los genes qnrS y blaVIM-11 en PARC y caracterizar el plásmido portador de qnrS.
Durante 2012 se recuperaron 38 PARC en un hospital de Buenos Aires. El tamizaje para detectar producción de metalo-beta-lactamasas (MBL) se llevó a cabo mediante sinergia de doble disco utilizando EDTA y carbapenems. El ADN plasmídico fue extraído utilizando fenol-cloroformo. Para determinar los alelos de los genes implicados en la síntesis de MBL y de PMQR, se llevó a cabo PCR-secuenciación. Para la detección de aac-(6′)-Ib-cr se realizó PCR-BseGI-RFLP. En aquellos aislamientos portadores de PMQR se secuenció el gen gyrA. Los grupos de incompatibilidad y sistemas de adicción fueron caracterizados por PCR. El plásmido portador de PMQR fue secuenciado completamente y curado manualmente.
De 38 aislamientos, 11 fueron productores de VIM (blaVIM-2 y blaVIM-11), mientras que uno contenía blaIMP-13. Si bien un aislamiento fue portador de blaVIM-11 y de qnrS1, dichos marcadores se encontraban en distintos plásmidos. El plásmido portador de qnrS1 (pP6qnrS1) presentó un tamaño de 117.945 pb y 154 secuencias codificantes (CDS); este correspondió al grupo de incompatibilidad IncR. Además de qnrS1, el plásmido portaba diversos marcadores de resistencia a aminoglucósidos. Aun cuando pP6qnrS1 no resultó conjugativo, presentó un oriT, de modo que posiblemente sea transferible.
Este es el primer informe acerca de PARC portadora de blaVIM-11 y de qnrS1 en simultáneo, además, es la primera descripción de un plásmido IncR en Argentina.
Pseudomonas aeruginosa is a ubiquitous non-fermenting Gram-negative rod, considered an opportunistic pathogen which can cause infections mainly in immunocompromised patients. Reports on this matter include hospital-acquired pneumonia, bacteremia, site infections in extensive burned areas, otitis and urinary tract infections2.
P. aeruginosa displays natural resistance to a wide set of antimicrobial agents including most β-lactams, thus narrowing the therapeutic options to a small group of antibiotics such as ceftazidime, cefepime, monobactams, carbapenems, fluoroquinolones, colistin and to a lesser extent, aminoglycosides. However, over the past years, resistance levels have increased by the acquisition of resistance mechanisms, rendering therapeutic alternatives useless. Mutations of the molecular target have become the main cause of quinolone resistance while impermeability-type mutants are mainly involved in resistance to aminoglycosides and carbapenems10. Even though inaccessibility to the molecular target is the main cause of carbapenem resistance in P. aeruginosa, metallo-β-lactamases (MBL) are a major determinant of transferable resistance. In Argentina MBL account for 65% of the enzymatic resistance to these antibiotics. In our region, blaVIM and blaIMP have been reported to be the most relevant MBL coding genes in P. aeruginosa, blaVIM-2 and blaIMP-13 being the most frequent variants of each gene16,18.
Plasmid-Mediated Quinolone Resistance (PMQR) genes confer diminished susceptibility to quinolones, facilitating the selection of higher-level resistance due to target mutation11. PMQR include genes coding for active efflux pumps (namely OqxAB and QepA), antibiotic modifying enzymes (aac-(6′)-Ib-cr) and a set of genes coding for target protective proteins belonging to the pentapeptide repeat family, generically named Qnr. The latter are the widest set of PMQR genes including 7 families (qnrA, qnrB, qnrC, qnrD, qnrS, qnrVC and qnrE) and their allelic variants11.
Broad host range resistance plasmids belonging to IncU and IncW groups have been reported in P. aeruginosa using PCR-based methods. However, in the case of plasmids native to the Pseudomonas genus, conjugation-based assays are more appropriate. Few documented cases of P. aeruginosa plasmids carrying qnr are available worldwide1,3,13,19. The aim of this study was to report the simultaneous presence of qnrS andblaVIM-11 in P. aeruginosa for the first time, and to fully characterize the qnrS-harboring plasmid.
Materials and methodsIsolates and antimicrobial susceptibilityA total of 38 carbapenem resistant P. aeruginosa isolates were recovered from a hospital in Buenos Aires city during 2012. Antimicrobial susceptibility tests were performed by the disk diffusion method according to CLSI guidelines, and those suggested by Sociedad Argentina de Bacteriología, Micología y Parasitología clínicas (SADEBAC) – AAM6,17.
Detection and characterization of MBL-coding genes.In order to detect the possible presence of MBLs, double disk synergy tests using imipenem (IPM), meropenem (MEM) and EDTA (1μmol) were carried out17. Plasmid DNA extraction was performed according to Kado & Liu. Screening for MBL genes was conducted by multiplex PCR according to Ellington et al.8 KPC-coding genes were investigated according to Bradford et al. in all isolates4. Several electroporation protocols and conjugation assays, using both E. coli DH5α and P. aeruginosa PAO-1 as recipient cells, were attempted9.
Simplex PCR reactions using blaVIM and blaIMP specific primers (Table 1) were used for further sequencing (Macrogen, Korea) and amplicon sequences were compared with the database using the NCBI BLASTn tool (www.blast.ncbi.nlm.nih.gov – 15/6/2019). The presence of MBL-coding genes in class 1 integrons was inferred combining 3́CS and 5́CS primers with either blaIMP or blaVIM forward and reverse primers (Table 1).
Primers used in this study.
| Primer name | Primer sequence (5′–3′) | Amplicon size (bp) |
|---|---|---|
| VIM START-F | ATGTTCAAACTTTTGAGTAAGT | 801 |
| VIM END-R | GCTACTCAACGACTGAGCG | |
| IMP-AF | GAAGGCGTTTATGTTCATACTT | 510 |
| IMP-AR | GTTTGCCTTACCATATTTGGA | |
| 5′CS | GGCATCCAAGCAGCAAG | – |
| 3′CS | AAGCAGACTTGACCTGA | – |
| qnrAF | ATTTCTCACGCCAGGATTTG | 516 |
| qnraR | GATCGGCAAAGGTTAGGTCA | |
| qnrSF | GCAAGTTCATTGAACAGGGT | 428 |
| qnrSR | TCTAAACCGTCGAGTTCGGCG | |
| qnrBF | GGMATHGAAAATCGCCACTG | 264 |
| qnrBR | TTTGCYGYYCGCCAGTCGAA | |
| qnrBCF | GTTRGCGAAAAAATTRACAG | 626 |
| qnrBCR | CCMATHAYMGCGATRCCAAG | |
| qnrCF | GGGTTGTACATTTATTGAATCG | 330 |
| qnrCR | CACCTACCCATTTATTTTCA | |
| qnrDF | CGAGATCAATTTACGGGGAATA | 582 |
| qnrDR | AACAAGCTGAAGCGCCTG | |
| qnrVCm-F | GAGYTKTATGGTTTAGAYCCTCG | 71 |
| qnrVCm-R | TGTTCYTGYTGCCACGARCA | |
| aac(6′)-IbF | CGATCTCATATCGTCGAGTGTT | 476 |
| aac(6′)-IbR | TTAGGCATCACTGCGTGTTC | |
| gyrAF-Pae | GAGCTGAAACAGTCCTATCTCGACT | 112 |
| gyrAR-Pae | TCATGGCATAAAGCACACGG |
PMQR markers were investigated using primers targeting the most common determinants (Table 1) and plasmid DNA as template. Additionally, the presence of aac-(6′)-Ib-cr was sought by PCR followed by digestion with BseGI. Amplicon sequences were identified using the NCBI BLASTn tool.
In those quinolone resistant isolates which were also positive for PMQR, the presence of mutations in the Quinolone-Resistance-Determining Region (QRDR) of gyrA was investigated on total DNA by PCR (Table 1) and sequencing (Macrogen, Korea). The QRDR sequence was compared with the same region of gyrA in PAO-1 (NC002516.2) using the BLASTn tool.
Plasmid characterizationPlasmid DNA harboring PMQR was extracted by a modification of the phenol-chloroform method proposed by Kado & Liu. Both, the PCR Based Replicon Typing scheme (PBRT) proposed by Carattoli et al.5 and addiction system detection proposed by Mnif et al.14 were performed.
Whole plasmid DNA was sequenced using Illumina Miseq technology. Fragment libraries were constructed by shotgun (NEB Ultra II) followed by 250bp paired-end sequencing; the reads were assembled using plasmid SPAdes V3.9 with default settings to include only contigs of more than 500 nucleotides. Genes were annotated using the RAST online tool and PROKKA software, contigs were thereafter manually curated and assembled. The complete plasmid sequence was compared with GenBank and transfer origin (oriT) databases (http://dnatools.eu/MOB/plasmid.html – 15/4/2019).
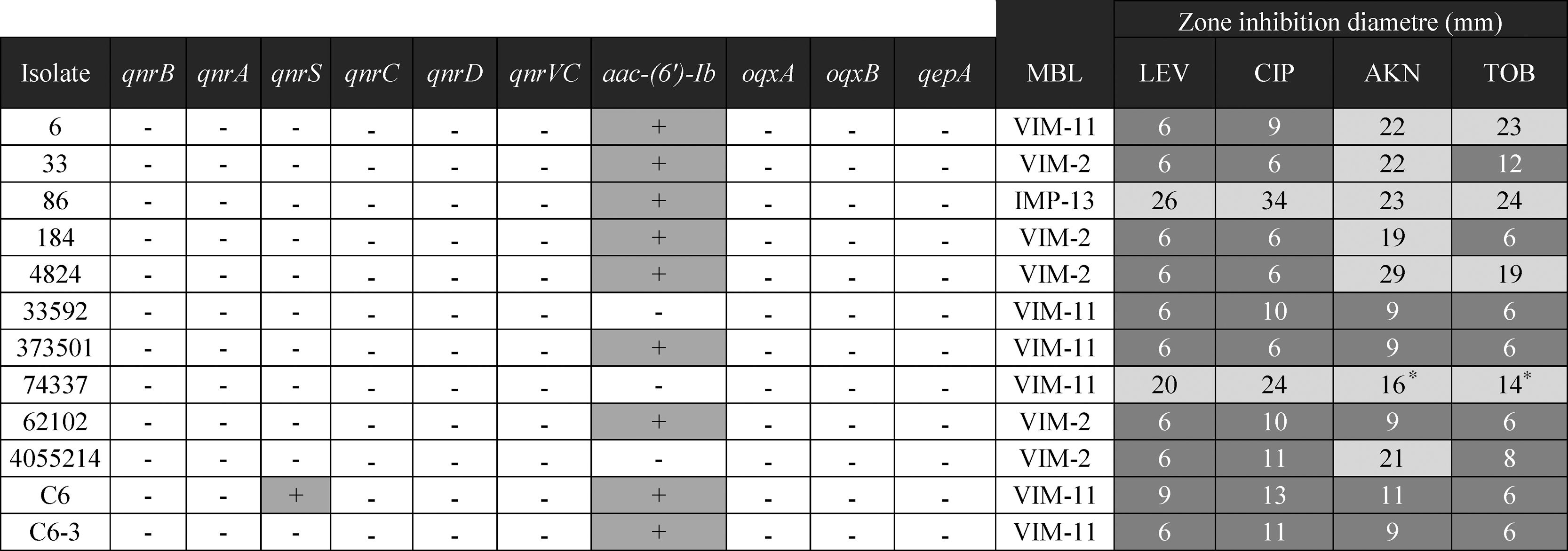
ResultsAntimicrobial susceptibility. Detection and characterization of resistance markersTwelve of the 38 (31%) carbapenem resistant P. aeruginosa isolates rendered positive in the double disk synergy test, suggesting the presence of an MBL. Ten of 12 isolates were also resistant to both ciprofloxacin and levofloxacin, and 8/12 to aminoglycosides (Table 2).
Using the MBL-multiplex PCR approach, blaVIM was detected in 11/12 isolates while blaIMP was detected in the remaining one. No KPC coding genes were detected in any of the 38 isolates. PCR amplification using specific primers and further sequencing identified blaVIM-2 (5), blaVIM-11 (6), and blaIMP-13 (1). Despite many attempts, none of the plasmids harboring blaVIM could be transferred. Both blaVIM and blaIMP were located as the only gene in the variable region of a class 1 integron.
One isolate, named PaeC6, rendered positive PMQR PCR results, the amplicon was further identified as qnrS1.PaeC6, also harbored blaVIM-11. No other PMQR determinant could be detected in any isolate. Even though aac-(6′)-Ib was detected in 10 isolates, none of them harbored the quinolone-modifying variant.
The gyrA QRDR of PaeC6 exhibited 2 substitutions, Thr83Ile and Ile89Leu, when compared to the same region in PAO-1.
Plasmid characterizationAll the plasmids were non-typeable by the PBRT scheme.
The plasmid harboring qnrS1 (pP6qnrS1) could not be transferred by electroporation. This plasmid was fully sequenced (A.N.: MH061383) and had a size of 117945bp with 154 CDS and an average G+C content of 51.55%. The replicon was contained in a region that spanned from the base 55069 to 64995, within this fragment the repB, parA and parB genes showed 100% identity with replicons of IncR plasmids either when using PlasmidFinder database or GenBank. This region also contained the partitioning site ParS and the UV reparation system umuCD. Near the 5′ end of this 9.9kb region, a resolvase coding gene (resD) was found, which could be potentially involved in the multimer resolution system of this plasmid. The replication regulator korC was located downstream the replicon region.
The addiction systems investigated by PCR according to Mnif et al. rendered negative results. However, in the plasmid sequence, the VapCB toxin-antitoxin system which belongs to the VagCD superfamily was detected at position 9500.
A tra region was found; nevertheless traS was missing and some of the remaining tra genes were either truncated or incomplete, thus explaining the failure to render transconjugants. An oriT sequence was detected between positions 100 478 and 100 873, displaying 100% identity with the same region of 3 different plasmids (NC004998, NC009837 and LT985322).
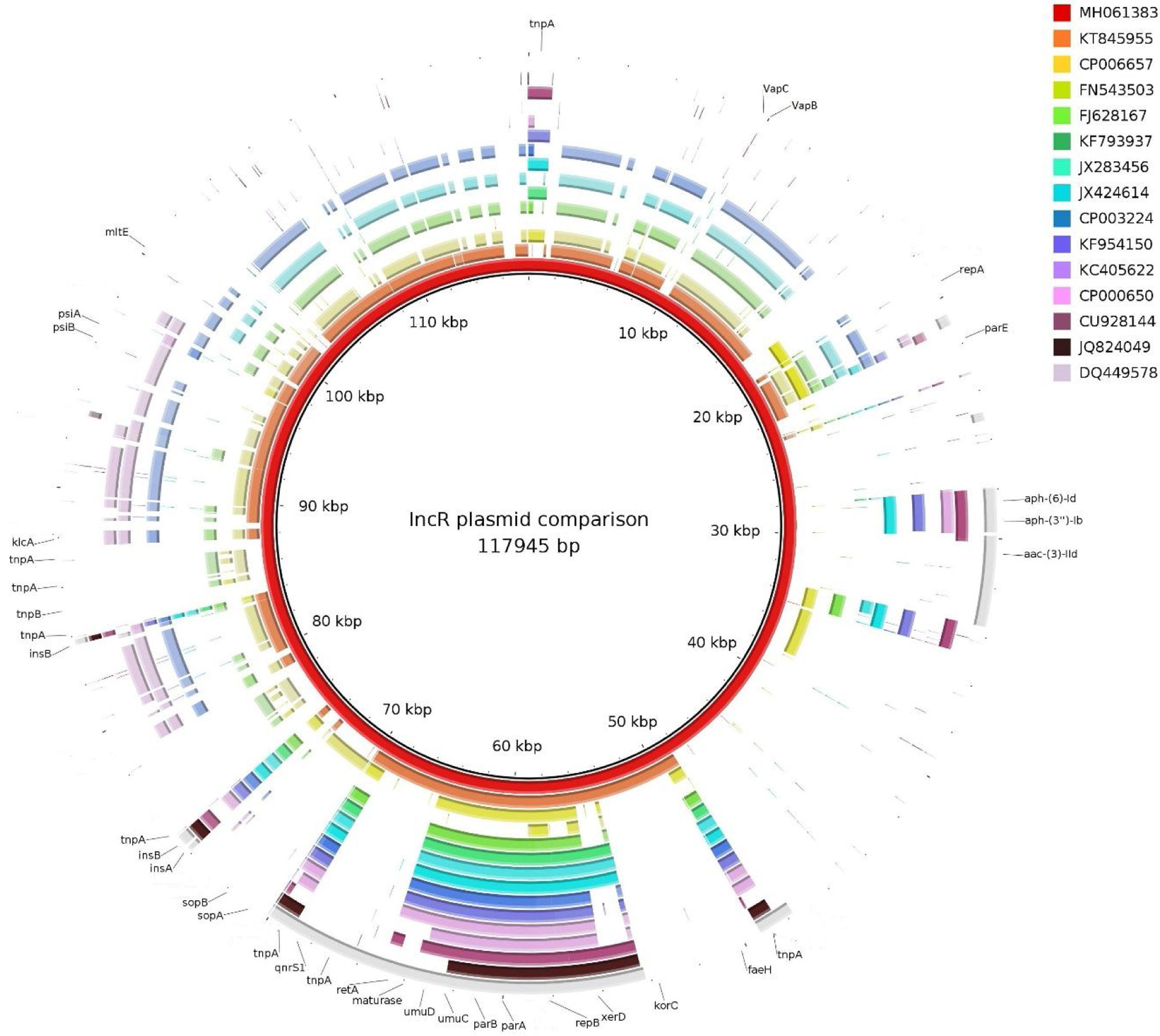
ISKpn19 was detected upstream of the replicon region, followed by qnrS1 and IS26. Furthermore, plasmid maintenance genes such as klcA, sopA, sopB were located following IS26 (Fig. 1). Additional aminoglycoside resistance genes were found along the plasmid such as aac-(3)-IId, aph-(6)-Id and aph-(3″)-Ib; however blaVIM-11 was not identified in this platform. Although the replicon of pP6qnrS1 corresponded to an IncR plasmid of P. aeruginosa, the overall backbone showed the strongest homology with that of pHNEP28_cfr from E. coli (AN: KT845955.1) (Fig. 2).
DiscussionEven though MBL production is not the prevalent carbapenem resistance mechanism in P. aeruginosa, their potential dissemination places it as the most clinically concerning. Reports on P. aeruginosa plasmids harboring MBL coding genes are common worldwide. The high plasticity of this species to acquire plasmid-borne resistance markers allows the horizontal genomic pool of P. aeruginosa to become a reservoir of a growing number of antibiotic resistance genes. The blaVIM and blaIMP alleles detected in this study are in accordance with those previously reported in Argentina12; nevertheless, to the best of our knowledge there are no previous reports of isolates carrying simultaneously MBL and PMQR determinants in P. aeruginosa in this country.
Although a wide set of plasmids harboring qnrS1 have been described in Enterobacteriales (IncColE, IncFI, IncHI, IncI1, IncL/M, IncN, IncX and IncR), the literature on PMQR in P. aeruginosa is scarce. There are descriptions of aac-(6′)-Ib-cr, qnrA1, qnrD, qnrS1 and qnrVC1; however only the last two markers were associated to β-lactamases, blaTEM-1 and blaVIM-2, respectively3,13. Although studies were carried out to detect PMQR in P. aeruginosa collected since 1990, the recent date of these reports may suggest that there is an ongoing increase in the mobile genetic pool in P. aeruginosa which could provide multiple resistance to the most clinically used antibiotics15.
The fluoroquinolone resistance observed in PaeC6 was allegedly the product of a pair of mutations in gyrA, one of which is known to generate the mentioned phenotype (Thr83Ile) while the other has not been described to date20. This should not, nevertheless, draw the attention out of the PMQR determinants found in this isolate since the presence of such markers is known to be a first step toward the selection of quinolone-resistant bacteria.
Just like the plasmids belonging to the IncR incompatibility group, pP6qnrS1 holds a conserved “replicon backbone” in which the genes necessary for the replication are located. In contrast, while most of the plasmids belonging to this incompatibility group show a set of genes related to plasmid maintenance immediately upstream of the replicon, the plasmid here described was found to harbor a PMQR marker, qnrS1, flanked by ISKpn19 and IS26 in this position.
In accordance with previous reports on IncR plasmids, pP6qnrS1 carries multiple IS and mobile elements, but also several resistance markers, mainly conferring resistance to aminoglycosides7. All IncR plasmids reported to date, including pP6qnrS1, are known to be non-transferable by conjugation, due either to a complete absence of some of the tra genes or to the presence of an abnormal truncated tra operon7.
IncR plasmid deposits are scarce in databases, none of them being from Argentina. Considering that plasmids belonging to this complex are known to be non-conjugative, the possible role of the emerging IncR complex in the spread of multidrug resistances is difficult to evaluate. However, the presence of an OriT in plasmid pP6qnrS1, makes it possible for this plasmid to be mobilizable.
Conflict of interestThe authors declare that they have no conflicts of interest.
This study was supported by PICT grants to DC, MR and GG (2015-2844, 2013-0858 and 2015-1925), UBACyT grants to MR and GG (20020150100174BA and 20020130100432BA), and by PIP grant 11220120100400CO to GG and MR.