The global epidemiology of fungal infections is changing. While overall, Candida albicans remains the most common pathogen; several institutions in Europe, Asia and South America have reported the rapid emergence to predominance of Candida parapsilosis. This mini-review examines the impact of gene deletions achieved in C. parapsilosis that have been published to date. The molecular approaches to gene disruption in C. parapsilosis and the molecularly characterized genes to date are reviewed. Similar to C. albicans, factors influencing virulence in C. parapsilosis include adherence, biofilm formation, lipid metabolism, and secretion of hydrolytic enzymes such as lipases, phospholipases and secreted aspartyl proteinases. Development of a targeted gene deletion method has enabled the identification of several unique aspects of C. parapsilosis genes that play a role in host–pathogen interactions – CpLIP1, CpLIP2, SAPP1a, SAPP1b, BCR1, RBT1, CpFAS2, OLE1, FIT-2.
This manuscript is part of the series of works presented at the “V International Workshop: Molecular genetic approaches to the study of human pathogenic fungi” (Oaxaca, Mexico, 2012).
La epidemiología mundial de las infecciones fúngicas está cambiando. Aunque Candida albicans sigue siendo el patógeno más común, varios centros en Europa, Asia y Sudamérica han descrito la rápida emergencia de Candida parapsilosis, que ha terminado por predominar. La presente revisión examina la influencia de las deleciones genéticas producidas en C. parapsilosis que se han publicado hasta la fecha. Se revisan las estrategias moleculares de la alteración de genes de C. parapsilosis y los genes caracterizados molecularmente hasta la fecha. Al igual que en C. albicans, los factores que influyen en la virulencia de C. parapsilosis incluyen la adherencia, formación de biopelículas, el metabolismo de lípidos y la secreción de enzimas hidrolíticas, como lipasas, fosfolipasas y aspartilproteinasas. El desarrollo de un método de deleción génica dirigido ha permitido la identificación de varios aspectos exclusivos de los genes de C. parapsilosis que participan en las interacciones huésped-patógeno-CpLIP1, CpLIP2, SAPP1a, SAPP1b, BCR1, RBT1, CpFAS2, OLE1, FIT-2.
Este artículo forma parte de una serie de estudios presentados en el «V International Workshop: Molecular genetic approaches to the study of human pathogenic fungi» (Oaxaca, México, 2012).
Since the late 1970s, fungal infections have increasingly become a significant cause of morbidity and mortality especially among hospitalized and immunosuppressed patients.69Candida species are the fourth most frequent causative agent of blood-stream infections, constituting 8–15% of hospital-acquired infections.87 In the United States, Candida albicans is the most common pathogen followed by Candida parapsilosis or Candida glabrata, depending on the study. However, C. parapsilosis has become the leading causative agent in some institutions located in Europe, Asia and South America, and it is the Candida species with the largest increase in incidence since 1990.1,6,10–12,20,36,48,52,54,70,74,81,87 Of all candidal isolates, C. parapsilosis accounts for 15.5% in North America, 16.3% in Europe and 23.4% in Latin America.81 In the US, it has been the third most common cause of neonatal sepsis.66
First isolated in 1928 from a stool specimen and thought to be non-pathogenic,2,86C. parapsilosis is now recognized as being fairly ubiquitous as it can be isolated from humans as a normal skin commensal as well as from domestic animals, insects, soil and marine environments.19,81,85 It is now especially well documented as a pathogen that arises from exogenous sources of infection in intravenous drug users and via medical instrumentation (e.g. catheters and hyperalimentation solutions).34,81 In particular, C. parapsilosis is recognized for its ability to cause invasive disease in patients without prior evidence of colonization via horizontal transmission through medical devices including catheters, parenteral nutrition solutions, and the hands of healthcare workers.81 Invasive disease occurs more often in immunocompromised patients, such as individuals with AIDS, cancer and in patients undergoing gastrointestinal surgical procedures.81 Other risk factors reported in studies include transplant receipt, antibiotic exposure, ophthalmic irrigating solutions and, especially, low birth weight in premature neonates.1,44,78,85,86 Clinical manifestations of C. parapsilosis include endocarditis, meningitis, peritonitis, arthritis, endophthalmitis, keratitis, otomycosis, onychomycosis, vulvovaginitis and urinary tract infections.81 Mortality rates attributed to C. parapsilosis range from 4% to 45%, with an average mortality rate of 28.5%.6,11,28,81 Biofilm producing isolates are associated with outbreaks38 and significantly higher mortality rates.82
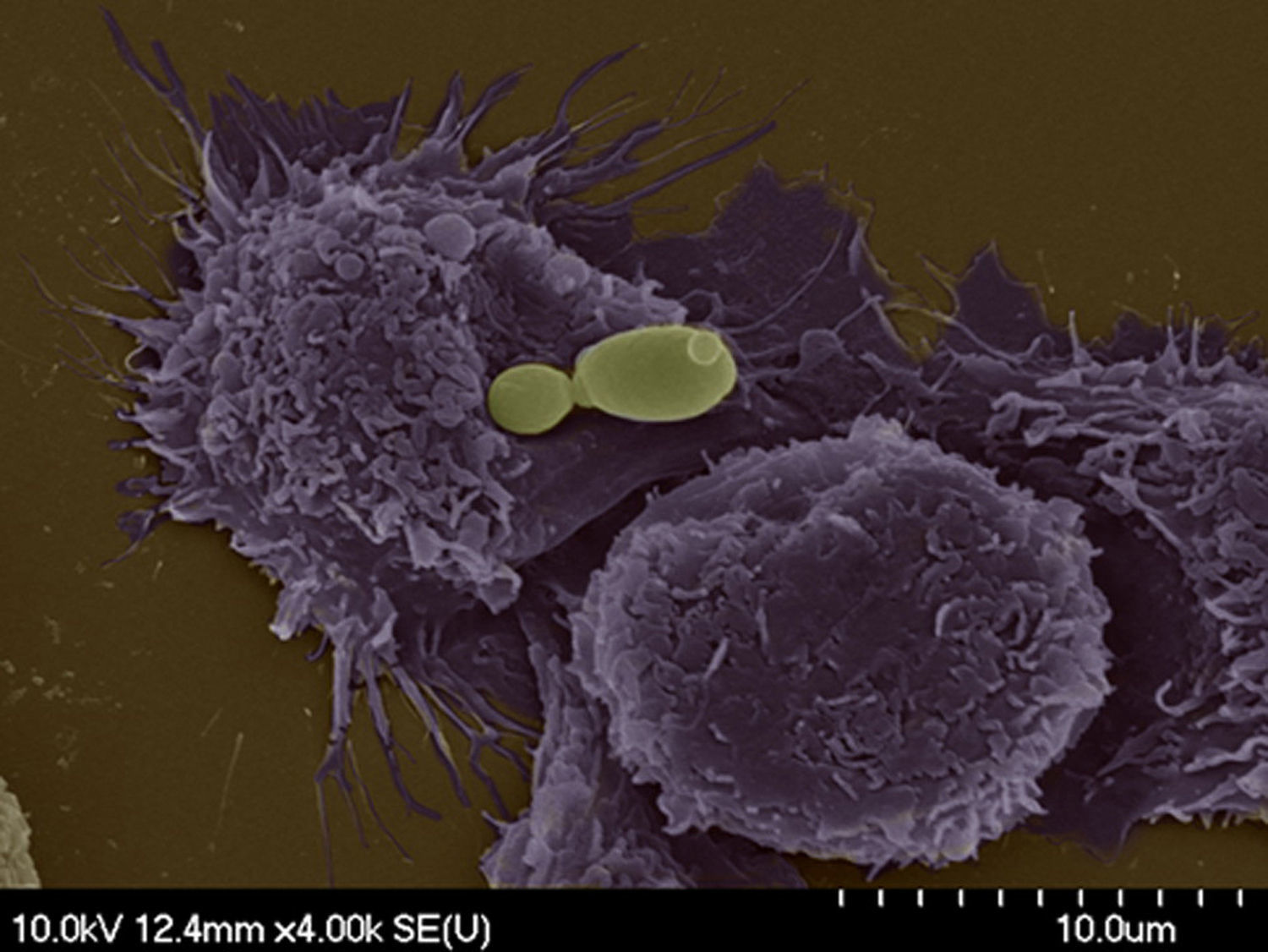
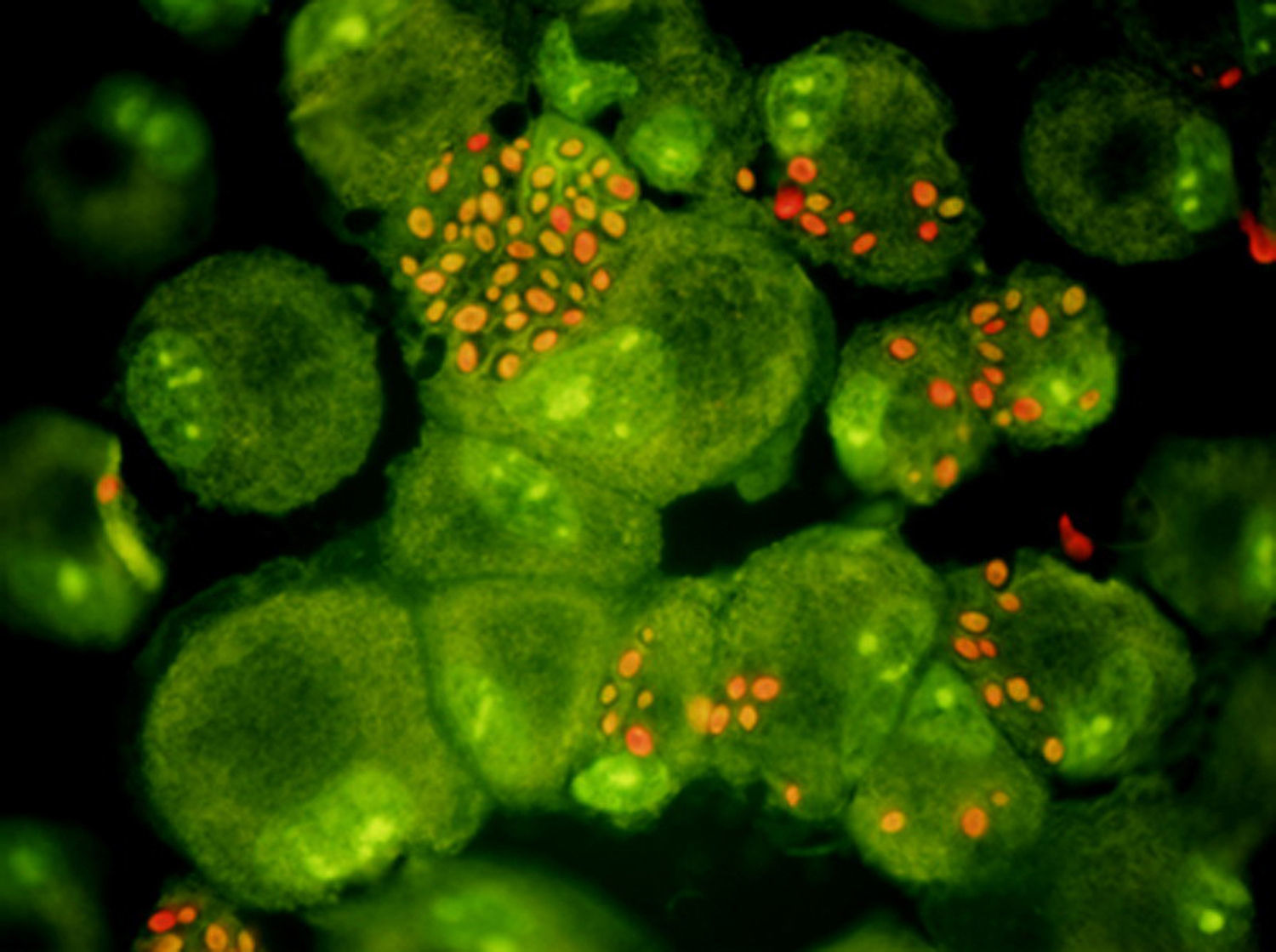
Determinants of virulence for candidal disease include adhesion capability to host surface, ability to switch morphology between yeast and filamentous growth, biofilm formation and secretion of extracellular hydrolytic enzymes such as lipases, phospholipases or secreted aspartyl proteinases.51,83 Conflicting data exist regarding phospholipase activity, with some studies demonstrating their presence in clinical isolates and others their absence.14,16,26,47 Nevertheless, its role in virulence bears consideration. The development of gene disruption methods to produce mutants has been a pivotal achievement in our capacity to gain insights into the interactions of C. parapsilosis with hosts and host effector cells (Figs. 1 and 2). This review will focus on the observations made on specific genes that have so far been characterized and shown to significantly influence these virulence traits.
C. parapsilosis has a diploid genome and does not have a described sexual cycle. Genetic analysis was initially limited by the availability of appropriate study tools. The first targeted gene disruption method was developed in 200723 based on previously established gene disruption protocols in C. albicans. The first targeted deletion in C. parapsilosis was the disruption of secreted lipases. This efficient gene deletion system was developed utilizing the repeated use of the dominant nourseoethricin marker (caSAT1) and subsequent deletion by FLP-mediated, site-specific recombination.23 Applying this technique, the lipase locus in C. parapsilosis containing the adjacent lipase genes CpLIP1, CpLIP2 were deleted and CPLIP2 reconstructed providing an understanding of the role of lipase activity in virulence.23
LipasesMicrobial extracellular lipases are virulence factors in a broad range of bacteria and fungi, including Candida species.81 So far, 10 lipase genes have been identified in C. albicans,32 and disruption, such as the deletion of LIP8, can significantly affect virulence.22 Only two lipase genes have been elucidated in C. parapsilosis – CpLIP1 and CpLIP2, of which only the latter has been demonstrated to code for an active protein.7,53 Utilizing the described targeted gene deletion method, disruption of these lipase genes provides evidence that they play a role in pathogenesis (i) by the decreased tissue damage seen in the presence of lipase inhibitors, (ii) the decreased ability of CpLIP1-CpLIP2 homozygous mutants to form complex biofilms, (iii) requirement for lipid-rich media, (iv) increased susceptibility to phagocytosis by macrophage-like cells, and (v) decreased virulence in comparison to wild-type C. parapsilosis yeast in infections of human oral epithelium or during murine intraperitoneal challenges.21,23 These observations hold significant clinical relevance since C. parapsilosis infections are particularly more commonly seen in patients receiving lipid-rich total parenteral nutrition and thus lipases may be a potential target for future antifungal agent development.81
Secreted aspartyl proteinase (Sap)Secreted aspartyl proteinase (Sap) genes have been demonstrated in most pathogenic Candida including C. albicans, Candida dubliniensis, Candida tropicalis and C. parapsilosis.25,45,49,89 However, they are notably absent in many non-pathogenic yeasts (e.g. Saccharomyces cerevisiae) suggesting their possible role in virulence.30 Sap isoenzymes have several functions such as (i) digestion of host proteins for provision of nitrogen sources, (ii) degradation of host cell surface structure and intracellular substances promoting tissue adhesion and invasion, and (iii) destruction of cells and molecules of the host immune system such as immunoglobulin G heavy chains, 2-macroglobulin, C3 protein, lactoglobulin, lactoperoxidase, collagen, and fibronectin71,77 thereby enabling evasion of antimicrobial activity.31 For example, vulvovaginal and skin C. albicans isolates exhibit higher in vitro Sap activity compared to blood isolates, which possibly explains the earlier clearance of fungemia in comparison to sustained skin infection in infection models.8,14,15,88 To date, ten SAP genes (Sap1p-Sap10p) have been identified in C. albicans.51 Their role in C. parapsilosis was unclear until the recent identification of 2 SAPP1 genes: SAPP1a and SAPP1b.30
The C. parapsilosis genome database (www.sanger.ac.uk/sequencing/candida/parapsilosis) was used to perform in silico analysis of the SAPP1 genes. A 2871 base pair-duplicated upstream (SAPP1a) and downstream (SAPP1b) regions were defined in the genome.30 All 4 alleles of the SAPP1 gene were deleted from wild-type (WT) C. parapsilosis.30 Separate homozygous ΔΔsapp1a, ΔΔsapp1b and a double-homozygous mutant ΔΔsapp1a-ΔΔsapp1b were generated.30 Subsequent experiments revealed that in an inducer medium, Sapp1p production was reduced by about 50% in the ΔΔsapp1a and ΔΔsapp1b mutants, but a similar effect was not observed for the SAPP2 gene. On the other hand, Sapp2p production was greater in the ΔΔsapp1a-ΔΔsapp1b mutant compared to wild-type yeast. This mutant was also hyper-susceptible to human serum, had decreased proteolytic activity, and decreased capacity to damage host-effector cells. Additionally, there was also greater phagocytosis and killing of the ΔΔsapp1a-ΔΔsapp1b yeasts by both human peripheral blood mononuclear cells (PBMCs) and PBMC-derived macrophages (PBMC-DM). After phagocytosis, the ΔΔsapp1a-ΔΔsapp1b yeasts induced more frequent phagolysosomal fusion, suggesting a role for Sapp1p in promoting intracellular survival.30 Both the lipase and proteinase genes play a major role in biofilm formation which is the hallmark of both bacterial and fungal organisms involved in device related infection. While the basic characteristics for biofilm formation may be similar among biofilm producing organisms, unique differences exist as well.
Biofilm formationColonization and infection due to C. parapsilosis are initiated by the organism's ability to adhere to host cells and tissues followed by the formation of biofilm on medical devices,81 which is accomplished in part by cell surface hydrophobicity67 and slime production.4 Compared to C. albicans, C. parapsilosis has a 20.6% greater avidity to buccal epithelial cells and 143.7% increased adhesiveness to acrylic material,67 although smaller studies have shown less significant differences.81 After adherence, biofilm formation is initiated by the establishment of cellular layers via cell-to-cell contact.75 In C. albicans, after adherence to tissues, the cells transform from yeast to hyphal forms, which appear to be a requirement for a structured biofilm.3,73 Once the mature biofilm is formed it is covered by an extracellular matrix that renders it less susceptible to antifungal medications. The transcriptional changes that occur during this process have been well studied in C. albicans, which reveal significant increases in the expression of genes that participate in glycolysis, amino acid and lipid metabolism.24,50C. parapsilosis and C. albicans differ in the nature of the biofilm formed. C. parapsilosis forms smaller and less complex structures,29,37 possibly because they do not produce hyphae. C. parapsilosis biofilms consist of yeast and pseudohyphal cells.17,23,37 Of note, pseudohyphal phenotypes can generate more biofilm and exhibit greater invasiveness into agar than strains in yeast forms.42 Similarities exist as well between the two species: biofilms of both are inhibited by exogenous farnesol42,76 and Bcr1 (Biofilm and Cell wall Regulator 1) is a major regulator for both species.17,60,61
BCR1In vivo rat catheter models have been utilized to describe the role of BCR1 in the development of biofilms.18BCR1 has been demonstrated to be a required fungal transcription factor for the formation of biofilms in both C. albicans and C. parapsilosis.17,60,61 Deletion of BCR1 in either species impairs the ability for biofilm formation.17,61 In C. albicans, BCR1 encodes genes targeting adhesins and cell-wall proteins (ALS1, ALS3, HWP1 and RBT5) indicating its role in the early adhesive stages of biofilm formation.60–63 The effect of Bcr1 on biofilm formation is also thought to be in part secondary to its influence on the expression of CFEM (Common in Fungal Extracellular Membranes) family of proteins. CFEMs were initially identified in Magnaportha grisea. They are similar to epidermal growth factor (EGF) domains that are found in extracellular membrane regions and contain an eight-cysteine domain.39,40 The role of CFEM is likely to act as cell surface receptors (adhesins).39 At least 5 CFEM members have been identified in C. albicans: PGA7, PGA10, RBT5, CSA1 and CSA2. Three of them, PGA10, RBT5 and CSA1, have been identified as vital for biofilm development.68
C. parapsilosis has seven CFEM members (CFEM1-CFEM7) including tandem duplicates of orthologs of C. albicans RBT5, PGA10 and CSA1.18CFEM1-CFEM4 are in tandem and syntenic with RBT5 and PGA-7, which in C. parapsilosis is postulated to have single gene duplications thereby forming CFEM1/CFEM2 and CFEM3/CFEM4. CFEM5-6 are orthologous with CSA1. An ortholog of CFEM7 has not been identified in the Candida clade and may possibly be specific to C. parapsilosis.18 Bcr1 exerts a different influence on the CFEM family in C. parapsilosis. In Bcr1d mutants, one member of each orthologous pair of CFEM2, CFEM3 and CFEM6 are down regulated, whereas expression of CFEM1, CFEM4, CFEM5 and CFEM7 is not affected.18 Interestingly, CFEM 2, CFEM3 or CFEM6 do not appear to be a requirement for the formation of biofilms in C. parapsilosis. CFEM2 and CFEM3 are necessary and CFEM6 is partially required for the heme utilization. Global transcriptional profiling of cells in an iron-depleted environment has shown the increased expression of 59 genes and decrease in 89 genes. Increased expression occurred in genes linked with cellular iron ion hemostasis and iron ion transport (FTH1, FRE9 and FRE10). Decreased expression was seen in heme containing and iron sulfur proteins (YHB1, SDH2 and ISA1) and all mitochondrial genes.18
RBT1When C. parapsilosis produces biofilms there is an upregulation of the genes involved in the glycolysis, fatty acid metabolism and ergosterol synthesis.75 These changes are similar to the ones observed when C. albicans cells are grown under hypoxic conditions.79 Although C. parapsilosis does not produce true hyphae, the genome includes a member of the hyphae producing gene family, Hwp1.76 One of the members in this family, the gene RBT1 is induced during the production of biofilms and under hypoxia.76 In C. albicans, RBT1 is induced during filamentation and mutants have been demonstrated to have decreased virulence in rabbit and mouse cornea models.5,33 In C. parapsilosis, RBT1 knockout isolates produce structurally much thinner biofilms than wild type while the heterozygous strains produce an intermediate thickness biofilm. This suggests that for full and appropriate biofilm development both of the RBT1 alleles are required.75
Lipid metabolismFatty acid formation is vital for the functioning of organisms in all kingdoms. They are building blocks of cell membranes that are products of cellular biosynthesis and require a complex enzyme system.58 So far, three major fatty acid synthesis systems have been identified. Eukaryotes and advanced prokaryotes (Mycobacterium, Nocardia and Corynebacterium) utilize Type I Fatty Acid synthesis system (FAS1), most bacteria utilize Type II, and parasites such as Trypanosma and Leishmania use FAS3.43 While similar enzymes are found in the three FAS systems, the organization of the encoding genes may significantly vary.58 In fungal organisms, production of essential fatty acids such as saturated and unsaturated fatty acids is critical for the generation and maintenance of cell membranes. Fatty acid synthase (FAS) and fatty acid desaturase (OLE) are important enzymes in this pathway.55
Fatty acid synthaseFungal FAS enzymes initiate the formation of a 2,6-MD heterodacemeric complex including subunits that are encoded by FAS1 and FAS2.58 These enzymes are therefore critical for normal yeast growth, and disruption of even a single gene can significantly alter the organisms’ physiological phenotype and virulence. Fas2 inhibition has been demonstrated to attenuate pathogenicity of Cryptococcus neoformans, C. parapsilosis and C. albicans.9,90,91
C. parapsilosis Fas2 (encoded by CpFAS2) heterozygous, homozygous and reconstituted Fas2 mutants have been generated.58CpFAS2 is required for growth in standard medium and gene expression was repressed in the presence of fatty acids.58 Further, up-regulation of CpFAS2 occurred when only glucose was made available as the carbon source. In comparison to a wild type yeast, CpFAS2 disruptants had a significant decrease in the concentration of unsaturated fatty acids.58CpFAS2 genes are also necessary for normal biofilm formation. Microarray studies demonstrate that CpFAS2 is upregulated during in vitro biofilm formation under hypoxic stress. Δfas2 strains were shown to have decreased capability for biofilm formation on polystyrene and silicone surfaces.58CpFAS2 also appears to play a vital role in helping the organism survive the fungicidal activity of monocytes such as neutrophils and macrophages. Intracellular survival of CpFAS2 disruptants was reduced by 40% when compared with wild type and heterozygous strains with a single FAS2 gene.58 This decrease in survival could likely be due to diminished membrane stability, thereby enhancing susceptibility to reactive oxygen species secreted by the macrophages. The Δfas2 strains demonstrate a leaky phenotype when grown under stress conditions. This action, in combination with defective fatty acid production, alters intracellular viability and proliferation of strains with CpFAS2 deletion.58
The Fas2 enzyme also appears to be critical for survival of C. parapsilosis in serum.55 This is a vital process in fungal pathogenesis as exposure to serum influences virulence traits such as filamentation and biofilm formation.59 Efficacy of antifungal drugs is decreased in serum, making the eradication of systemic infections difficult.65CpFAS2 disruptants are hypersensitive to serum and induce cell death.55 Cell survival also appears to be influenced by the presence of glucose, which causes mitochondria-dependent cell death. Mechanisms of glucose toxicity in Δfas2 strains are yet unclear, as glucose is usually a preferred carbon source for normal yeast growth. Toxicity may potentially be secondary to uncontrolled metabolism of glucose by the mutant cells, thereby creating an imbalance of cellular contents and subsequent triggering of a cell death response,55 especially via energy-requiring apoptosis pathways, overproduction of ROS, nuclear fragmentation and cell shrinkage.27,84
Fatty acid desaturase (OLE1)In addition to glucotoxicity, elevated lipid content is thought to be detrimental to normal yeast growth.57 In C. parapsilosis, exposure to high glucose induces formation of lipid droplets (LD); it is considered a possible mechanism through which yeast cells survive gluco- and lipo-toxicity.57 Both the CpFAS2 and fatty acid desaturase genes (OLE1) appear to play a role. Disruption of either of these genes inhibits LD formation from glucose.57 Moreover, OLE1 inhibition resulted in gluco/lipotoxicity of log phage yeast cells. While survival of wild type yeasts was decreased after 2 days of incubation, Δole1 mutants died within a few hours of incubation, indicating its role in glucose detoxification.57 In addition to glucose, OLE1 deletion strains are also hypersusceptible to fructose, galactose, and mannose.
FIT2Lipid droplets (LDs) are cytoplasmic compartments that contain triacylglycerides (TAGs) that serve as fatty acid reservoirs46 and lipid precursors of fatty acids (FA) and diacyl glycerol (DAG) for membrane lipids such as glycerophospholipids and sphingolipids.13,41 TAGs and steryl esters are major components of LDs and utilize fatty acyl CoA as a common substrate.72 Akin to mammalian cells, LDs may significantly influence the functioning of pathogenic fungi. In C. parapsilosis, formation of LDs is necessary for normal cell growth and virulence.56 A family of proteins involved in LD formation – fat storage-inducing transmembrane proteins 1 and 2 (FIT1 and FIT2) – has recently been demonstrated in both humans and mice.35,56 In mammalian cells, FIT2 orthologs enable compartmentalization of TAG in LDs and depleting FIT2 transcripts in adipocytes decreased LD formation.35 The FIT2 proteins are primarily localized to the endoplasmic reticulum and are responsible for lipid partitioning rather than lipid synthesis.35 Effects of FIT2 on LD accumulation are therefore likely downstream in comparison to the DGA1 pathway.64,80
In C. parapsilosis, disruption of FIT2 impairs LD formation, alters the lipidome and attenuates virulence.56 Both in vitro and in vivo studies demonstrated that heterozygous strains with deletion of one FIT2 allele did not decrease TAG formation or LD size. However, deletion of both FIT2 alleles significantly decreased TAG content (39%) and LD accumulation.56 Alterations were also noted in other lipid species with FIT2 deletion. While a 25% reduction was observed in fatty acids (FFA), unsaturated fatty oleic (4.63%) and linoleic acids (12.31%), elevations were noted in steryl esters (135%), diacylglycerols/sterols (96%) and phospholipids (112%), palmitic (1.5%) and stearic acids (15%).56 However, FIT2 deletion did not alter susceptibility to standard antifungal drugs.
ConclusionsThe epidemiology of microorganisms that impact human morbidity and mortality is constantly evolving. Globally, there is increased life expectancy, improved and more frequent access to the healthcare system, rising use of immunosuppressants, immunomodulators and biologics, and increased utilization of indwelling devices such as catheters and cardiovascular devices. Microbes that have the ability to evade the host immune system, produce biofilms and develop resistance to available treatment options have and will significantly impact morbidity and mortality. This is well exemplified by the globally increasing prevalence of C. parapsilosis infections. A significant work has been accomplished in identifying the genetic factors that determine the organism's virulence, and increased efforts are urgently needed to counter this global threat.
Conflict of interestsThe authors have no conflicts of interest.
AG was supported in part by OTKA NF 84006, NN100374 (ERA-Net PathoGenomics Program) and an EMBO Installation Grant. JN was supported in part by an award from the Irma T. Hirschl/Monique Weill-Caulier Trust.