Selecting the best embryos to transfer is essential in any in vitro fertilization treatment. For years, embryo morphology supported this selection. Now, the field is seeking more accurate and non-invasive methods for selection. Recent studies show that embryos that do not result in successful pregnancy exhibit different metabolomic profiles from those embryos that result in successful pregnancy, opening a new possibility in the field. Metabolomic profiling and analysis of the cell-free DNA released by the embryo to the culture media are the most promising techniques for embryo selection developed so far.
Seleccionar el mejor embrión para transferir es esencial en cualquier tratamiento de fecundación in vitro. Durante años, se ha utilizado la morfología embrionaria para llevar a cabo esta selección. Hoy en día se están investigando métodos de selección más precisos y no invasivos. Estudios recientes muestran que embriones que no han dado lugar a un embarazo presentan un perfil metabolómico que difiere de aquellos que sí acaban en un embarazo, abriendo nuevas posibilidades en el campo. El perfil metabolómico y el análisis del DNA libre circulante liberado por el embrión al medio de cultivo son, actualmente, las técnicas más prometedoras para la selección embrionaria.
Infertility is one of the most prevalent global health conditions, affecting approximately 15–20% of individuals of reproductive age worldwide. The major causes of infertility in women include anovulation, anatomic problems, and endometriosis; in men, sperm disorders are the critical causes (Asampille et al., 2020). Many of these challenges are overcome by assisted reproductive technology (ART). Among the treatment modalities offered to infertile couples, in vitro fertilization (IVF) is associated with the highest success rates (SART, 2018).
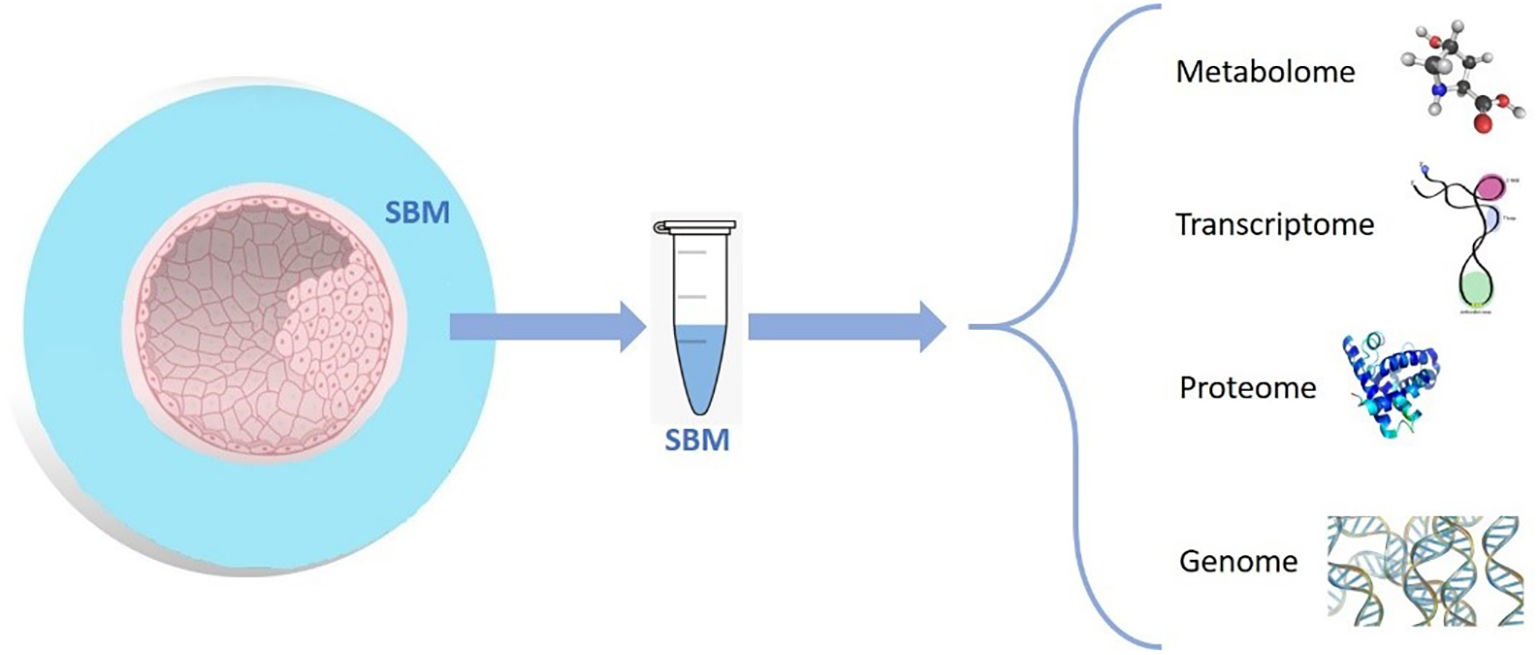
For decades, IVF commonly resulted in multiple gestations; the incidence of this outcome has gradually decreased due to recent advances in embryo culture and cryopreservation and implementation of high-quality single embryo transfers (sET) (Gardner et al., 2015). Conventional selection of embryos for transfer requires morphological evaluation, which is considered safe, precise, and straightforward (Cummins et al., 1986). However, this static mode of evaluation entails a series of drawbacks, such as subjectivity of the evaluator, need for frequent assessment during embryo development, and culture stability (Rocha et al., 2016). These limitations are partially overcome with time-lapse systems that enable not only analysis of embryo morphology but also collection of information on the dynamic changes during the preimplantation period (Conaghan, 2014; Chavez-Badiola et al., 2020; Meseguer et al., 2011). Despite the progress made by implementing such systems, the field continues to seek adjunctive technologies to estimate the reproductive potential of an embryo. These technologies combine the proteomic, metabolomic, and genomic (Fig. 1) characteristics of the embryo with the conventional morphologic criteria to select the embryo most likely to produce a pregnancy (Sigalos, Triantafyllidou and Vlahos, 2016).
Scheme of the different molecules that can be found in the SBM. Metabolome (lipids, amino acids, carbohydrates, and nucleotides); transcriptome (mRNAs, small non-coding RNAs, and long non-coding RNAs); proteome (structural proteins, non-structural proteins, functional proteins, and enzymes); and genome (embryo cfDNA to determine the embryo ploidy).
Because embryos undergo metabolic shifts during development, assessment of the metabolome—the sum of metabolic analytes—is the most widely investigated non-invasive OMICs approach in embryo assessment (Uyar and Seli, 2014). Yet, its efficiency is not fully known. This review presents the basic aspects of preimplantation embryo metabolism, outlining the current leading technologies to assess embryo metabolic states. We also present a detailed literature review demonstrating correlation between the culture media metabolome and clinical outcomes. Finally, the advantages and limitations of metabolomics research in IVF are discussed, emphasizing potential future applications that may move the field toward non-invasive embryo selection.
MetabolomicsMetabolomics in systems biologyMetabolomics is a rapidly evolving field that uses advanced analytical chemistry techniques in conjunction with sophisticated statistical methods to comprehensively characterise the metabolome (German, Hammock and Watkins, 2005). The metabolome is commonly defined as the complete collection of metabolites (<1 kDa), or small molecule chemicals, found in a given organelle, cell, organ, biofluid, or organism (Pasikanti, Ho and Chan, 2008). These small molecule chemicals include endogenous compounds such as lipids, amino acids, short peptides, nucleic acids, sugars, alcohols, or organic acids (Wishart, 2019). Metabolite synthesis is encoded by the genome, providing valuable information about genotype–phenotype relationships and genotype–environment interactions(Demain, 1980). Compared with the human genome (~20,300 genes) or the human proteome (>620,000 protein species) the human metabolome is estimated to include ~3,000 metabolites (Seli, Robert and Sirard, 2010). This relatively small set of metabolites can be efficiently profiled, making metabolomics a powerful tool in biomedical research (Uyar and Seli, 2014). Traditionally, metabolomic research analyses biological fluids that are not limiting in volume, such as blood or urine. However, embryo metabolomic usually implies analyzing limited volumes and a small concentration and number of metabolites. Metabolic profiles can be non-invasively measured in spent culture blastocyst media (SBM) to identify what the embryo has taken up and produced during culture, thus providing an approximate assessment of embryo quality. Likewise, metabolomics enables identification of biomarkers related to maternal and perinatal complications and contributes to our understanding of the physiopathology of the most complex and prevalent diseases, maternal/foetal infections, and other severe maternal morbidities (Fanos et al., 2013; Dessì, Marincola and Fanos, 2015; Kamath-Rayne et al., 2014; Li et al., 2016).
Analysis of metabolomics dataSeveral analytical techniques are used to study embryo metabolism for IVF. These include nuclear magnetic resonance (NMR) spectroscopy (Seli et al., 2008; Pudakalakatti et al., 2013); mass spectrometry (MS) (Cortezzi et al., 2013), which can be coupled with separation methods such as gas chromatography (GC-MS), liquid chromatography (LC-MS or HPLC-MS), or capillary electrophoresis (CE-MS); and near-infrared (NIR) (Seli et al., 2007) and Raman spectroscopies(Ding et al., 2017).
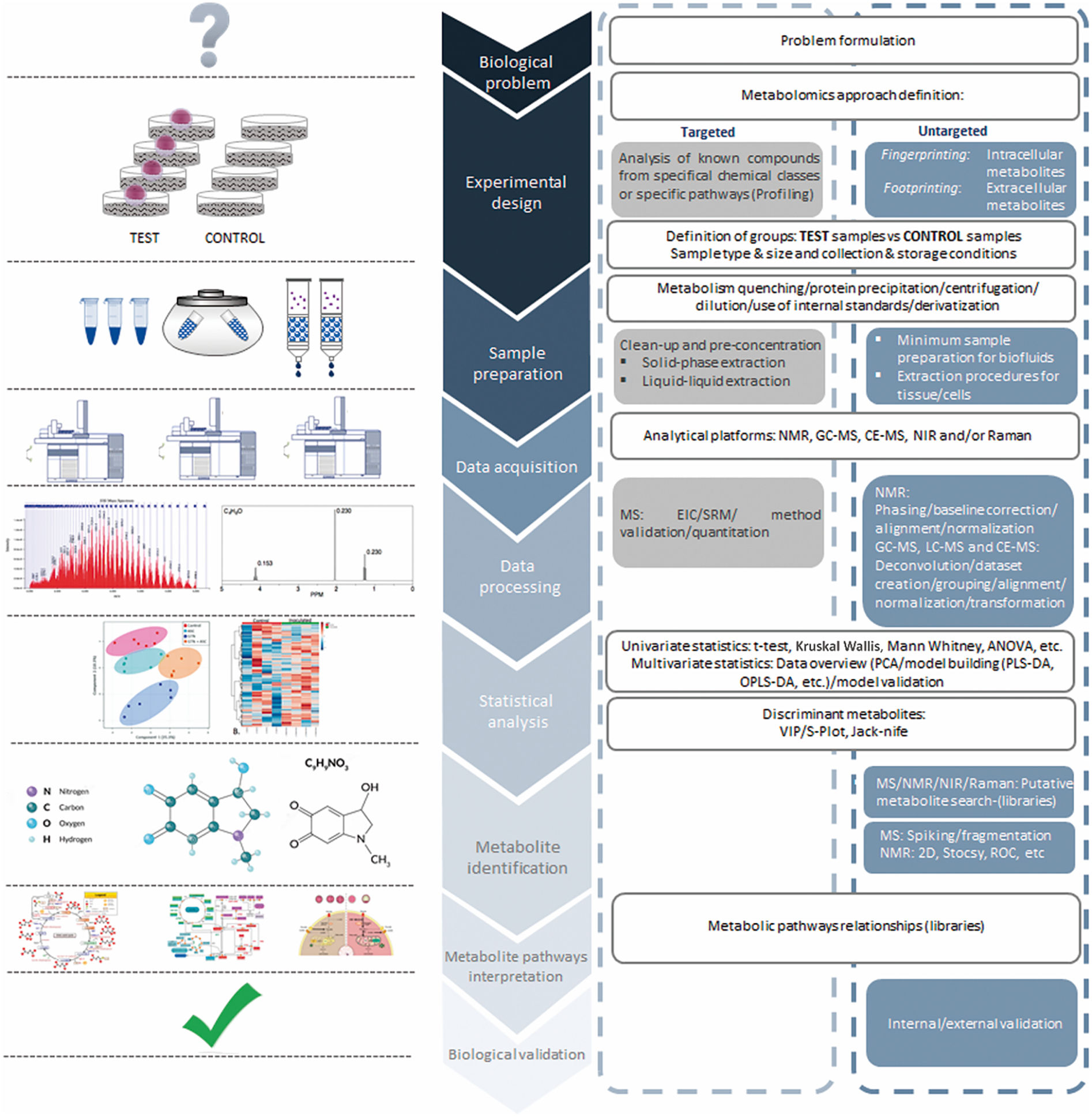
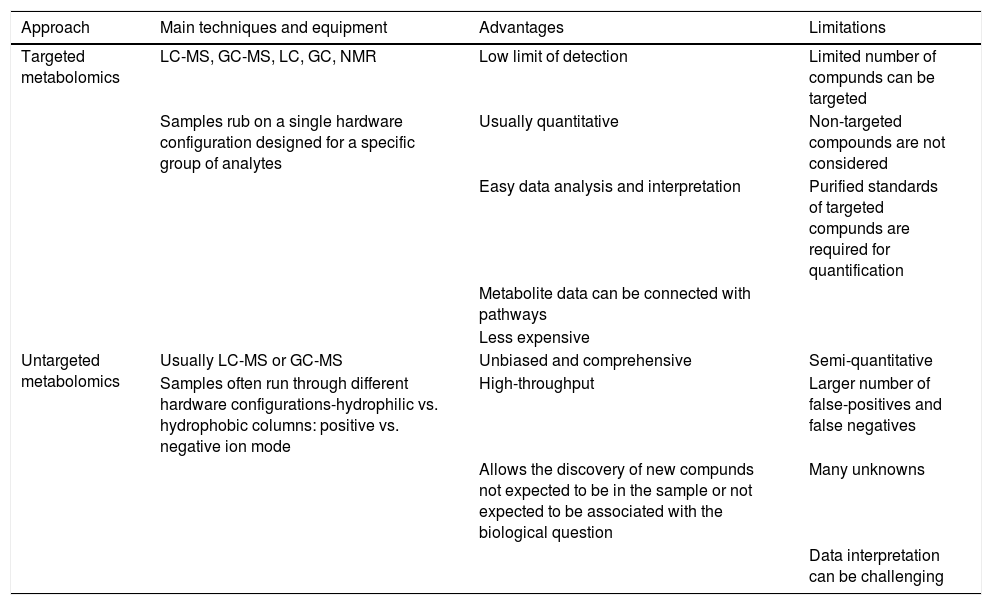
Generally, there are four types of metabolic experiments that can be selected depending on their purpose and the instrumental capabilities of the laboratory performing the study: 1) targeted metabolomics, 2) untargeted metabolomics, 3) fluxomics, and 4) metabolite imaging (Wishart, 2019). Among them, targeted and untargeted metabolomics approaches are the most common workflows carried out in the context of ART (Table 1). Targeted metabolomics is preferable for hypothesis testing and biomarker detection. In the targeted approach, the chemical properties of the investigated compounds are known, and sample preparation can be tailored to reduce matrix effects and interference from accompanying compounds. The targeted approach is also responsive to high-throughput, kit-based systems using NMR, LC-MS, or GC-MS equipment and appropriate software (Roberts et al., 2012; Xia et al., 2013). This approach has been used for over three decades for the advancement of ART by quantification of one or several known metabolites used by the embryo, such as glucose, lactate, amino acids, or ammonium.
Comparison of targeted and untargeted analysis strategies in metabolomics.
| Approach | Main techniques and equipment | Advantages | Limitations |
|---|---|---|---|
| Targeted metabolomics | LC-MS, GC-MS, LC, GC, NMR | Low limit of detection | Limited number of compunds can be targeted |
| Samples rub on a single hardware configuration designed for a specific group of analytes | Usually quantitative | Non-targeted compounds are not considered | |
| Easy data analysis and interpretation | Purified standards of targeted compunds are required for quantification | ||
| Metabolite data can be connected with pathways | |||
| Less expensive | |||
| Untargeted metabolomics | Usually LC-MS or GC-MS | Unbiased and comprehensive | Semi-quantitative |
| Samples often run through different hardware configurations-hydrophilic vs. hydrophobic columns: positive vs. negative ion mode | High-throughput | Larger number of false-positives and false negatives | |
| Allows the discovery of new compunds not expected to be in the sample or not expected to be associated with the biological question | Many unknowns | ||
| Data interpretation can be challenging |
On the other hand, untargeted metabolomics is ideal for metabolite discovery and hypothesis generation. It generally uses LC-MS, GC-MS, or CE-MS to characterise as many metabolites or putative metabolites as possible (Patti, Yanes and Siuzdak, 2012; Schrimpe-Rutledge et al., 2016). While untargeted metabolomics is often very labour-intensive, it has led, among other milestones, to the creation of a relative “embryo viability score” intended to reflect embryo developmental potential (Vergouw et al., 2008).
A critical step in the metabolomics workflow (Fig. 2) is accurate and efficient analysis of high-dimensional complex metabolomics data. Such data can be analysed using a wide range of statistical methods (Shulaev, 2006). For metabolic assessment of embryo viability, normalisation constitutes an indispensable pre-processing step due to systematic variations in the resulting spectra data derived from the multistage experimental setting. In this context, the main source of non-biological variations in embryo assessment may be attributed to the culture environment. Spectral profiles of SBM may be normalised to those of blank samples to eliminate the possible impact of variations in culture conditions. Following normalisation, predictive models can be developed using machine learning algorithms intended to predict implantation potentials of individual embryos (Alpaydin, 2010).
Importantly, these metabolomic-based embryo selection models need to be carried out using multicentre data and validated in different centres to ensure model robustness in case of centre-specific variations. Likewise, culture media metabolomics can be integrated with data from other OMICs platforms to provide an extensive knowledge base for functional genomics research in reproductive sciences (Mehrotra and Mendes, 2006).
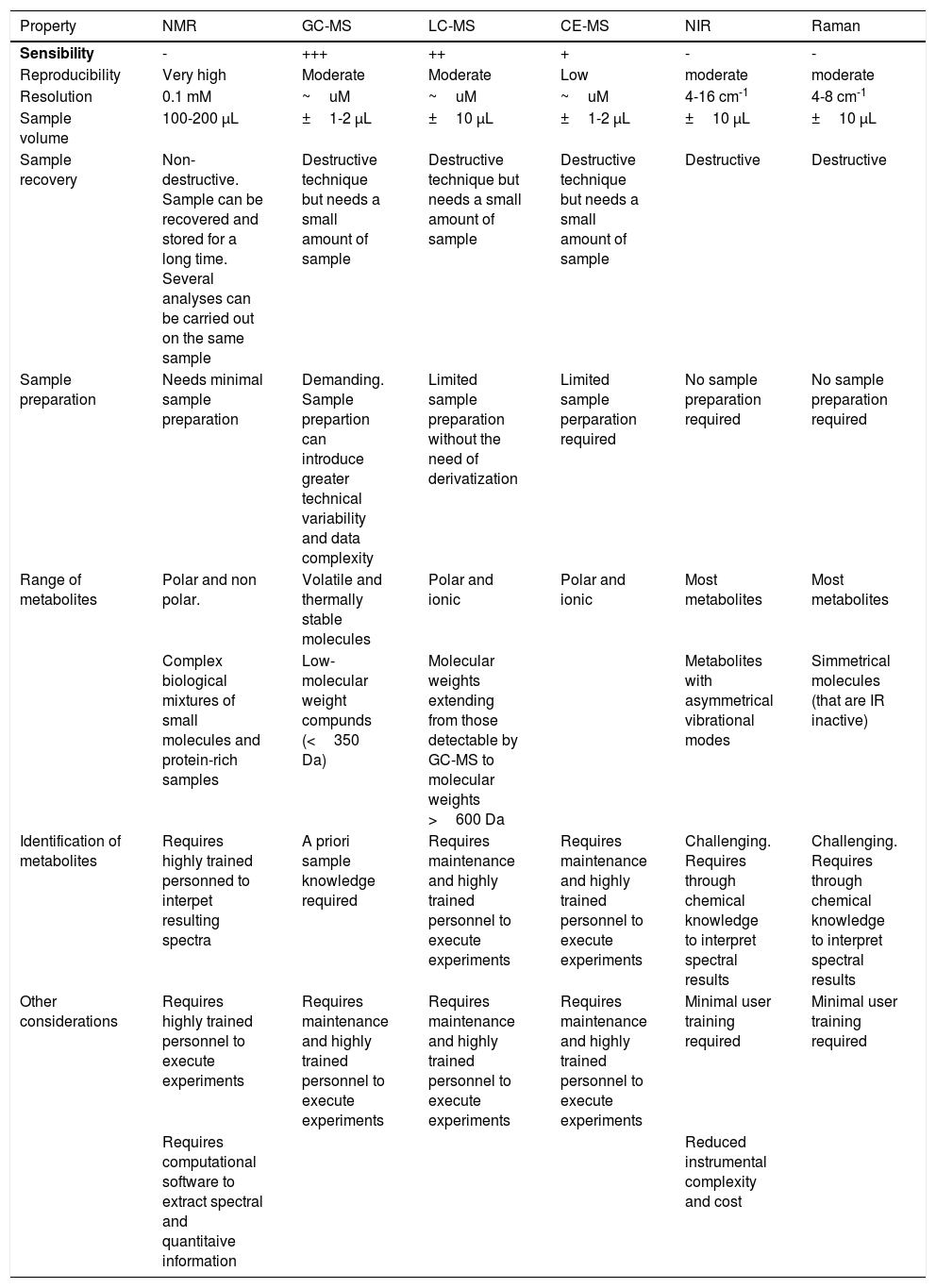
Analytical technologiesMany different customisations are available for metabolomic analytical methods, allowing better identification of a specific range of molecules or metabolites with different solubilities. Therefore, the advantages and disadvantages of each method should be considered according to the experimental objectives (Table 2). This section briefly describes the analytical platforms that are commonly used for metabolomics research in IVF and related data analysis methods.
Comparison of analytical techniques in metabolomic profiling.
| Property | NMR | GC-MS | LC-MS | CE-MS | NIR | Raman |
|---|---|---|---|---|---|---|
| Sensibility | - | +++ | ++ | + | - | - |
| Reproducibility | Very high | Moderate | Moderate | Low | moderate | moderate |
| Resolution | 0.1 mM | ~uM | ~uM | ~uM | 4-16 cm-1 | 4-8 cm-1 |
| Sample volume | 100-200 μL | ±1-2 μL | ±10 μL | ±1-2 μL | ±10 μL | ±10 μL |
| Sample recovery | Non-destructive. Sample can be recovered and stored for a long time. Several analyses can be carried out on the same sample | Destructive technique but needs a small amount of sample | Destructive technique but needs a small amount of sample | Destructive technique but needs a small amount of sample | Destructive | Destructive |
| Sample preparation | Needs minimal sample preparation | Demanding. Sample prepartion can introduce greater technical variability and data complexity | Limited sample preparation without the need of derivatization | Limited sample perparation required | No sample preparation required | No sample preparation required |
| Range of metabolites | Polar and non polar. | Volatile and thermally stable molecules | Polar and ionic | Polar and ionic | Most metabolites | Most metabolites |
| Complex biological mixtures of small molecules and protein-rich samples | Low-molecular weight compunds (<350 Da) | Molecular weights extending from those detectable by GC-MS to molecular weights >600 Da | Metabolites with asymmetrical vibrational modes | Simmetrical molecules (that are IR inactive) | ||
| Identification of metabolites | Requires highly trained personned to interpet resulting spectra | A priori sample knowledge required | Requires maintenance and highly trained personnel to execute experiments | Requires maintenance and highly trained personnel to execute experiments | Challenging. Requires through chemical knowledge to interpret spectral results | Challenging. Requires through chemical knowledge to interpret spectral results |
| Other considerations | Requires highly trained personnel to execute experiments | Requires maintenance and highly trained personnel to execute experiments | Requires maintenance and highly trained personnel to execute experiments | Requires maintenance and highly trained personnel to execute experiments | Minimal user training required | Minimal user training required |
| Requires computational software to extract spectral and quantitaive information | Reduced instrumental complexity and cost |
NMR: Nuclear magnetic resonance; GC-MS: Gas-chromatography-mass spectrometry; LC-MS: Liquid chromatography-mass spectrometry; CE-MS: Capillary electrophoresis mass spectrometry; NIR: Near infrared spectroscopy
Nuclear magnetic resonance (NMR)-based metabolomic analysis resulted largely from work by Nicholson et al. (Nicholson et al., 2002; Nicholson et al., 1984). This non-optical spectroscopy technique takes advantage of the magnetic moment of specific atomic nuclei with an external magnetic field to provide information about metabolites that contain elements with non-zero magnetic moments. As a non-destructive analytical tool, NMR is efficiently used for biomarker analysis by enabling detection and quantification of specific metabolites within a biological fluid or tissue (Emwas, 2015). Despite having lower sensibility, NMR offers a series of advantages that make it one of the first-choice analytical techniques for metabolic-screening: 1) NMR spectra are highly reproducible (Wong, 2014), 2) the non-destructive nature of the technique allows usage of the sample for other purposes (Mishkovsky and Frydman, 2009), 3) NMR spectroscopy is intrinsically quantitative, thus enabling precise quantification of precursors and products (Wishart, 2008; Barding, Salditos and Larive, 2012; Truong, Yoon and Shanks, 2014), and 4) it can be used indistinctly for both targeted and untargeted analysis of metabolic flux in vivo and in vitro (Nargund et al., 2013). Limitations of NMR include the requirement for large amounts of sample, higher costs, and lack of sensitivity for low-abundance targets. Likewise, NMR experiments can also be time-costly, ranging from less than a minute to a few hours per measurement.
Mass spectrometryMass spectrometry (MS) operates through three steps: (i) ion formation, (ii) separation of ions according to their mass-to-charge ratios (m/z), and (iii) detection of separated ions (Dunn and Ellis, 2005). MS enables simultaneous characterisation of several hundreds of metabolites with higher sensitivity than NMR approaches, as MS has the capability to detect metabolites at micromolar concentrations. The sensibility and specificity of MS are further enhanced when coupled with chromatography or electrophoresis-based separation techniques (Pasikanti, Ho and Chan, 2008).
Gas-chromatography-mass spectrometryGas-chromatography-mass spectrometry (GC-MS) generally serves as a versatile analytical platform due to its robustness, excellent separation capability, selectivity, sensitivity, and reproducibility. Other advantages include ease of use and its ability to provide insight into compound identification. However, a fundamental limitation of GC-MS is that it can only separate and identify low molecular weight (50-600 Da) and volatile compounds (Garcia and Barbas, 2011). For detection of polar, thermolabile, and non-volatile compounds, chemical derivatisation is required prior to analysis. Such an approach introduces more significant technical variability and complexity to the data, as a single metabolite can produce multiple derivatised peaks (Dunn and Ellis, 2005).
Liquid chromatography-mass spectrometryLiquid chromatography-mass spectrometry (LC-MS) approaches are commonly used and favoured for metabolomic analysis due to their high throughput, soft ionisation and good coverage of metabolites (phospholipids, proteins, amino acids, glycosides, and sugars) (Dunn and Ellis, 2005). Unlike GC-MS, LC-MS can work with a wide range of molecular weights, from low molecular weights to molecular weights >600 Da. Also, the mobile phase is liquid, allowing increased coverage compared to GC since all compounds may not reach the volatility level required by GC. These conditions simplify sample preparation and make LC-MS ideal for metabolomic analysis of biological fluids (Zhou et al., 2012).
Capillary electrophoresis mass spectrometryCapillary electrophoresis mass spectrometry (CE-MS) offers fast and high-resolution separation of highly polar and charged analytes from small injection volumes (few μL) (Monton and Soga, 2007). Coupled to MS, it represents a powerful analytical technique capable of analysing a wide range of analytes from inorganic compounds to large proteins (Sastre Toraño, Ramautar and de Jong, 2019). Unlike other MS techniques, CE-MS does not require rigorous sample pre-treatment, making it especially well-suited for working with small amounts of material (Monton and Soga, 2007). Despite its advantages, CE-MS remains generally underrepresented as an analytical approach in metabolomics. Limited use may be attributed to poor sensitivity, migration time variability, and lack of standardisation.
Vibrational spectroscopyThe use of NMR and chromatographic MS techniques in the clinical setting is limited by their cost, lack of reproducibility, and practicality as a commercial bench-top product (Botros, Sakkas and Seli, 2008). Alternatively, vibrational spectroscopy (NIR and Raman spectroscopy) offers a series of advantages from a technical perspective (Seli et al., 2007). The instrumentation employed with these techniques is easier to handle than that required for NMR or MS. Also, the instruments are highly stable over time and can be maintained and operated by minimally trained users (Dunn and Ellis, 2005).
There is no single best analytical technique for all IVF metabolomics approaches. Therefore, choosing one will depend on the metabolite class of interest, required sensitivity, dynamic range of measurements, sample size, sample-specific pre-treatment, and the relative time and cost-efficiency of the method (Uyar and Seli, 2014).
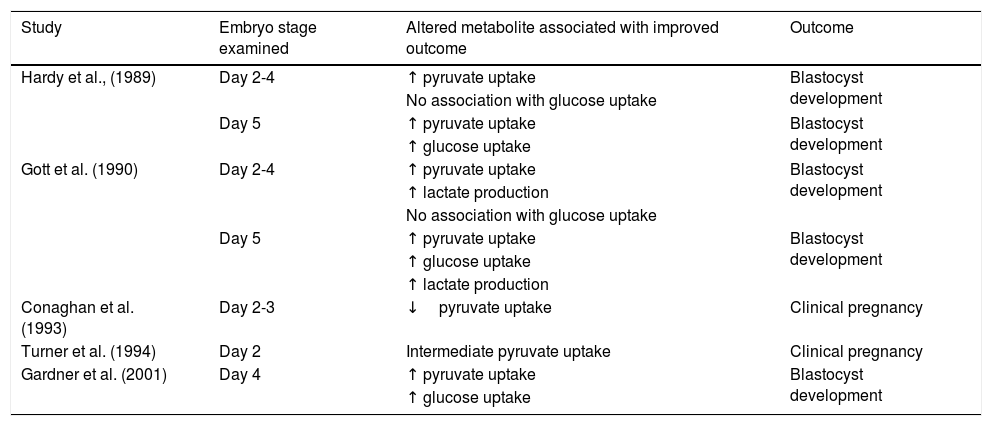
Embryo metabolism: What is currently known?The backbone of our current understanding of embryo metabolism comes from early studies of carbohydrate metabolism in mice (Brinster, 1965; Biggers, Whittingham and Donahue, 1967). In general, early-cleavage-stage embryos primarily use pyruvate, lactate, and amino acids. In contrast, later-stage embryos switch to a reliance on glucose metabolism via glycolysis as the blastocyst forms and expands (Gardner and Leese, 1987; Gardner et al., 2001). The most suitable-quality embryos show elevated glucose uptake compared to poorer quality ones (Renard, Philippon and Menezo, 1980; Gardner and Leese, 1987). A similar phenomenon occurs in human embryos, where those that arrive at the blastocyst stage and correlate with morphologic grade have higher glucose consumption (Gardner et al., 2001) (Table 3). Several studies sought to identify a possible marker of growth potential regarding pyruvate metabolism (Hardy and Spanos, 2002; Gott et al., 1990; Conaghan et al., 1993a; Conaghan et al., 1993b; Turner et al., 1994; Gardner et al., 2001; Hardy et al., 1989); however, whether pyruvate uptake is predictive of embryo development and viability remains inconclusive. While some studies report higher pyruvate uptake in embryos that develop to the blastocyst stage (Hardy et al., 1989; Gott et al., 1990), others demonstrate an inverse relationship between pyruvate uptake by 2–8-cell embryos and embryo viability and pregnancy (Conaghan et al., 1993a; Conaghan et al., 1993b) (Table 3).
Pyruvate, lactate, and glucose metabolism as predictors of embryo development and viability.
| Study | Embryo stage examined | Altered metabolite associated with improved outcome | Outcome |
|---|---|---|---|
| Hardy et al., (1989) | Day 2-4 | ↑ pyruvate uptake | Blastocyst development |
| No association with glucose uptake | |||
| Day 5 | ↑ pyruvate uptake | Blastocyst development | |
| ↑ glucose uptake | |||
| Gott et al. (1990) | Day 2-4 | ↑ pyruvate uptake | Blastocyst development |
| ↑ lactate production | |||
| No association with glucose uptake | |||
| Day 5 | ↑ pyruvate uptake | Blastocyst development | |
| ↑ glucose uptake | |||
| ↑ lactate production | |||
| Conaghan et al. (1993) | Day 2-3 | ↓pyruvate uptake | Clinical pregnancy |
| Turner et al. (1994) | Day 2 | Intermediate pyruvate uptake | Clinical pregnancy |
| Gardner et al. (2001) | Day 4 | ↑ pyruvate uptake | Blastocyst development |
| ↑ glucose uptake |
Amino acids also play an essential role in preimplantation embryo metabolism. Uptake and production of amino acids are correlated with outcomes such as DNA damage, ploidy, embryo sex, and embryo quality (Sturmey et al., 2009; Picton et al., 2010). In this context, decreased culture medium levels of glycine and leucine and increased levels of asparagine correlate with clinical pregnancy and live birth (Brison et al., 2004). Also, higher glutamate levels are associated with a better prognosis (Seli et al., 2007). Most importantly, a low amino acid turnover is linked with better development. For this mode of action, the “quiet embryo hypothesis” (Leese, 2002; Leese et al., 2007; Leese et al., 2008; Baumann et al., 2007) maintains that viable embryos have reduced metabolism because they are able to respond to cellular stressors more efficiently. This is based on the knowledge that embryo metabolism needs to increase to meet the energetic demands required for coping with cellular stressors. In support of this hypothesis, several studies report a connection between reduced metabolic activity and developmental potential or increased metabolic activity and cellular stress. Further, monitoring patterns of oxygen consumption in human embryos in culture for up to 72 hours may be informative of embryo quality (Tejera et al., 2012).
Despite the efforts in targeted metabolite analysis, there is not yet a single biomarker of embryo quality since the metabolic picture is far too complicated to be explained by a single metabolic imbalance (Bracewell-Milnes et al., 2017). Therefore, the strategy has moved towards global embryo metabolomic assessment, known as metabolomic profiling.
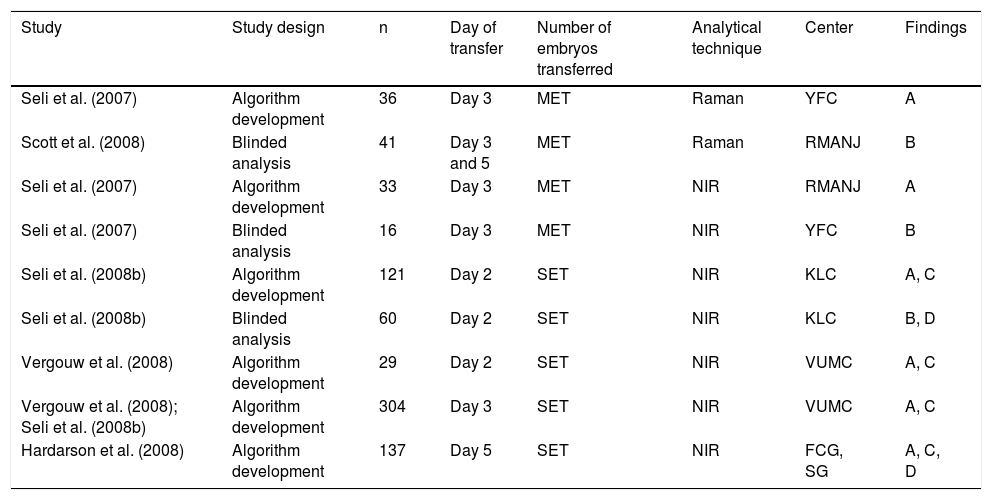
Can metabolomic profiles be used to identify the suitability of preimplantation embryos?Several validation studies were published during 2007 and 2008, using a multivariate analysis approach to study the metabolomic profiles of the SBM from embryos used for sET and multiple embryo transfer (mET). The goal was to assess whether embryo metabolomic status and clinical outcome could be linked (Botros, Sakkas and Seli, 2008).
The first metabolomics study in embryos was published in 2007 by Seli et al. (2007). They used NIR and Raman spectroscopy to obtain metabolomic profiles from the SBM of 69 Day-3 embryos from 30 patients with known outcomes. The authors compared the metabolic profiles of embryos that implanted and resulted in a live birth with those of embryos that did not implant to develop a viability score (or viability index) algorithm. Interestingly, Raman spectroscopic analysis of SBM of embryos with proven reproductive potential demonstrated higher viability indexes (0.5886 + 0.2222) than those that failed to implant (0.3264 + 0.2884; P>0.01). The viability score algorithm was able to identify implantation/pregnancy potential with a specificity of 76.5% and a sensitivity of 85.7%. In 2008, Scott et al. (2008) used the algorithm developed by Seli et al. (2008) to analyse 44 blinded SBM samples from Day-3 (n=35, from 14 patients) and Day-5 (n=9, from 5 patients) embryos from 19 patients in different clinics. The media and volumes used to culture the embryos differed among the IVF clinics. They found that Day-3 embryos showed significantly higher viability indexes than Day-5 embryos (0.71 + 0.07 vs 0.51 + 0.13, P<0.0001). These discrepancies could reflect the significant differences in embryo metabolism at different stages of development. The sensitivity and specificity were 85.7%. The authors concluded that there is a strong association between SBM metabolomic profiles and clinical outcomes.
Subsequent studies had larger sample sizes and aimed to validate the use of metabolomic profiling from SBM collected upon sET and mET (Table 4). In 2008, Seli et al. (2008) used proton NMR to analyse SBM of 34 Day-3 embryos from 18 patients with known pregnancy outcomes (0% or 100% sustained implantation rates). As in previous studies, the mean viability index of embryos with proven reproductive potential was significantly larger compared with those embryos that failed to implant (0.6201 + 0.1619 vs. 0.3799 + 0.2660). Sensitivity and specificity using proton NMR to identify implantation/pregnancy were each 88.2%.
Studies of the non-invasive metabolomic profile of SBM to assess embryo viability in IVF cycles.
| Study | Study design | n | Day of transfer | Number of embryos transferred | Analytical technique | Center | Findings |
|---|---|---|---|---|---|---|---|
| Seli et al. (2007) | Algorithm development | 36 | Day 3 | MET | Raman | YFC | A |
| Scott et al. (2008) | Blinded analysis | 41 | Day 3 and 5 | MET | Raman | RMANJ | B |
| Seli et al. (2007) | Algorithm development | 33 | Day 3 | MET | NIR | RMANJ | A |
| Seli et al. (2007) | Blinded analysis | 16 | Day 3 | MET | NIR | YFC | B |
| Seli et al. (2008b) | Algorithm development | 121 | Day 2 | SET | NIR | KLC | A, C |
| Seli et al. (2008b) | Blinded analysis | 60 | Day 2 | SET | NIR | KLC | B, D |
| Vergouw et al. (2008) | Algorithm development | 29 | Day 2 | SET | NIR | VUMC | A, C |
| Vergouw et al. (2008); Seli et al. (2008b) | Algorithm development | 304 | Day 3 | SET | NIR | VUMC | A, C |
| Hardarson et al. (2008) | Algorithm development | 137 | Day 5 | SET | NIR | FCG, SG | A, C, D |
SET: single embryo transfer. MET: multiple embryo transfer. YFC: Yale Fertility Center, USA. RMNAJ: Reproductive Medicine Associates New Jersey, USA. VUCM: Vrije Universiteit Medical Center, The Netherlands. KLC: Kato Ladies Clinic, Japan. FCG: Fertilitets Centrum, Sweden. SG: Shady Grove Reproductive Science Center, USA.
A: Mean viability score of embryos that implanted and resulted in fetal cardiac activity or live birth was significantly higher compared with the mean viability score of embryos that failed to implant. B: Spectroscopy analysis by an investigator blinded to pregnancy outcome using a previously established regression algorithm demonstrated that the mean viability score of embryos that resulted in a pregnancy was higher compared with embryos that failed to implant. C: Study showed the metabolomic profile of embryo culture media to be independent of morphology. D: A positive correlation was detected between increasing viability scores and the potential individual embryos to result in a pregnancy.
Interestingly, the more studies were performed, the more consistent the findings were: embryos with higher mean viability scores resulted in more pregnancies with foetal heartbeat (Botros, Sakkas and Seli, 2008).
These studies suggested that IVF embryos with high potential could alter the SBM differently than embryos that do not result in pregnancy. Moreover, these detectable differences could inform selection of the best embryos to transfer. Currently, metabolomic profiles are not widely used to select embryos for transfer, but rapid evolution of this field yielded significant new information. In particular, results are available for two randomised controlled trials (RCTs) (Vergouw et al., 2012). In both studies, patients were divided into control and treatment groups. In the control group, embryos were transferred according to morphology; in the treatment group, embryos were transferred according to morphology and metabolomic profiling (using NIR). The results among both groups were not statistically different, potentially because the embryos in the treatment group were also selected using morphology. Vergouw et al. (2012) reported the worst live birth rates in the treatment group. Hence, further studies are necessary to validate the proposed algorithms in different types and volumes of media and to design RCTs that only study the metabolomic profiles when transferring the embryos. Liang et al. (2019) compared the ploidy status vs. the Raman spectra (54 euploid and 33 aneuploid embryos), suggesting that ploidy status could be related to changes in the metabolomic profile. Nevertheless, the number of samples with results was low; hence, more studies are needed before translating metabolomic profiles to the clinical setting.
Another embryo feature to bear in mind is the chromosomal content of the embryos. Nearly half of the embryos produced in IVF treatments have an incorrect number of chromosomes (Ata et al., 2012). These anomalies are mostly de novo (Franasiak et al., 2014; Rubio et al., 2019) and are related to advanced maternal age and other factors of the couple (such as altered karyotype, recurrent implantation failure). When transferred, most aneuploid embryos end up in miscarriage in the first trimester (Sugiura-Ogasawara et al., 2012; Kung et al., 2015); or result in a birth with a chromosomopathy.
To avoid these negative outcomes, the embryo transferred in an IVF cycle must contain the correct number of chromosomes (Rubio et al., 2019). Chromosomal abnormalities may involve either loss or gain of a whole chromosome, known as uniform aneuploidy, and/or small deletions or duplications (del/dup) of a fragment of a chromosome, known as partial or segmental aneuploidy (García-Pascual et al., 2020). Diagnosing these anomalies in the embryo allows clinicians to avoid transfer of aneuploid embryos.
To determine the chromosomal make-up of preimplantation embryos, programs widely use preimplantation genetic testing for aneuploidies, known as PGT-A (García-Pascual et al., 2020). PGT-A is performed in a trophectoderm biopsy of the embryo; hence, it is an invasive procedure. Developing non-invasive strategies to select the best embryo to transfer could offer important options in the field. One such strategy is time-lapse microscopy complemented by special predictive algorithms. Another is analysis of the cell-free DNA (cfDNA) released by the embryos to the SBM (Vendrell and Escribà, 2021). Embryos go from a relatively inactive metabolism at ovulation to a fast metabolism at implantation; thus, how metabolomic and proteomic markers in the SBM correlate with embryo viability is of interest (Zmuidinaite, Sharara and Iles, 2021). In fact, preimplantation embryos show an important autonomy in vitro, producing their own trophic factors and achieving a dialog with the endometrium, which allows successful implantation and invasion (Kane, Morgan and Coonan, 1997; Navarrete Santos et al., 2008).
Conclusions and future perspectivesNearly 50% of the embryos generated in IVF treatments are aneuploid (Munné, 2006). These embryos will fail in implantation, result in miscarriage, or produce a newborn with a syndrome. Hence, both invasive and non-invasive procedures to test which embryos have the best opportunities to implant and develop into healthy babies are important to supporting good IVF outcomes. Embryo morphology and, more recently, morphokinetics—a combination of morphology and embryo kinetics—are the first non-invasive approaches used to select the embryos with the best developing potential. Nevertheless, time-lapse monitoring has not been proven to discern between chromosomally normal and abnormal embryos (Del Carmen Nogales et al., 2017).
Other strategies are in development. There are differences in depletion and/or excretion of metabolites among embryos (Motiei et al., 2020), so euploid embryos may have different metabolomic profiles than those with aneuploidies. The latest studies suggest that chromatography would be the best technique to study the embryo metabolome (Mádr et al., 2015; Motiei et al., 2020). Nonetheless, the overall depletion/appearance of metabolites may vary significantly between studies due to differences in the composition of the culture media, the developmental stage of the embryos, variability in the measurement techniques, and viability of the embryos (Motiei et al., 2020). Several groups have tried to develop “metabolomic profiles” to select embryos with higher implantation potential. Nevertheless, to date, neither technique is reported as successful in the clinical setting.
One promising strategy is the analysis of the cell-free DNA (cfDNA) released by the embryo into the culture media. First described by Shamonki et al. in 2006 (Shamonki et al., 2016), analysis of cfDNA is now starting to be used in the clinical setting (Rubio et al., 2020) to rank the embryos with more probability of being euploid. To provide robust evidence, more studies are needed. Rubio et al. are carrying out an RCT to test whether transferring an embryo based on analysis of cfDNA in the medium is better than transfer according to morphology.
In conclusion, several non-invasive approaches provide an opportunity to guide selection of the best embryos to transfer. However, more studies are needed to support the use of metabolomics as a biomarker for embryo transfer.