Preimplantation genetic testing is implemented worldwide as an effective tool to avoid transmission of single gene disorders and/or chromosome abnormalities. This approach requires the obtention of representative samples from embryos in order to infer its genetic status. Nowadays, the embryo biopsy is the first-choice method for embryo sampling. Biopsy procedures are safe and widely performed in the clinical routine. However, this intervention is invasive and requires trained personnel and investment in specific equipment. Recently, new sampling methods have been suggested under the term of “non-invasive”. These approaches are based on the existence of cell-free DNA into the embryo or its environment. An increasing number of studies suggest the collection of fluid from embryos or spent culture medium to obtain cell-free DNA to assess the genetic condition of preimplantation embryos avoiding embryo biopsy. The reliability of this attractive idea needs to be confirmed and validated. In this sense, this work offers a deep review of data published to date. Several methods of DNA detection, quantification and amplification have been tested and different protocols and culture systems, with or without additional embryo manipulations, are being investigated. In general terms, an enormous variability among published results is noteworthy. Central aspects as DNA detection rates, contamination with extraembryonic DNA and concordance between results (cell-free DNA versus embryo samples) are centring proofs-of-concept and validation experiments. However, basic questions as the biological origin and representativeness of cell-free DNA are pending to be answered. Solved, unsolved and serious limitations of the new approaches are discussed. A final reflection respecting the state of the technique is offered.
Las pruebas genéticas de preimplantación se aplican en todo el mundo como una herramienta eficaz para evitar la transmisión de trastornos de un solo gen o de anomalías cromosómicas. Este enfoque requiere la obtención de muestras representativas de los embriones para inferir su estado genético. Hoy en día, la biopsia embrionaria es el método de primera elección para la toma de muestras de embriones. Los procedimientos de biopsia son seguros y se realizan ampliamente en la rutina clínica. Sin embargo, esta intervención es invasiva y requiere personal capacitado e inversión en equipos específicos. Recientemente, se han propuesto nuevos métodos de muestreo bajo el término de «no invasivos». Estos enfoques se basan en la existencia de ADN libre de células en el embrión o en su entorno. Un número cada vez mayor de estudios proponen la recogida de líquido de los embriones o del medio de cultivo empleado para obtener ADN libre de células con el fin de evaluar el estado genético de los embriones de preimplantación y evitar la biopsia embrionaria. La fiabilidad de esta atractiva idea debe ser confirmada y validada. En este sentido, este trabajo ofrece una profunda revisión de los datos publicados hasta la fecha. Se han probado varios métodos de detección, cuantificación y amplificación del ADN y se están investigando diferentes protocolos y sistemas de cultivo, con o sin manipulaciones adicionales del embrión. En términos generales, destaca una enorme variabilidad entre los resultados publicados. Aspectos centrales como las tasas de detección de ADN, la contaminación con ADN extraembrionario y la concordancia entre los resultados (ADN libre de células frente a muestras de embriones) están centrando las pruebas de concepto y los experimentos de validación. Sin embargo, cuestiones básicas como el origen biológico y la representatividad del ADN libre de células están pendientes de respuesta. Se discuten los problemas resueltos, los no resueltos y las graves limitaciones de los nuevos enfoques. Se ofrece una reflexión final respecto al estado de la técnica.
Preimplantation genetic testing (PGT) of human embryos generated in vitro is globally implemented in the clinical routine depending on countries’ legal framework. In the past decades, different modalities of PGT have been developed in order to avoid the transmission of monogenic disorders (PGT-M), unbalanced structural chromosome rearrangements (PGT-SR), chromosome aneuploidies (PGT-A), selection of HLA-compatible embryos (PGT-HLA) or the combination of all together (Carvalho et al., 2020). Sample collection is one of the limiting factors to obtain sufficient and representative embryonic DNA for genetic testing. From the PGT framework, sample collection has changed throughout embryo development, from the zygote, cleavage to blastocyst stage. Nowadays, the biopsy of a few (five to ten) trophectoderm cells from the blastocyst is considered the first-choice method of sampling in the majority of IVF (in vitro fertilization) laboratories. It is a safe procedure ensuring accuracy, reliability and reproducibility. It has also show not impact on subsequent embryo viability and reproductive potential (Cimadomo et al., 2016). However, biopsy protocols are invasive, technically challenging, requiring specific equipment and trained personnel. In addition, an accurate biopsy is a crucial step to maximize the results of genetic testing (Kokkali et al., 2020), always avoiding to compromise the blastocyst implantation potential. This critical step has activated the research of new forms of embryo sampling and new strategies are being evaluated in the frame of “non-invasive” approaches. Non-invasive PGT (ni-PGT) methodologies aim to obtain representative DNA from the embryo without contact (or by a minimal intervention) with the embryo structures.
The first attempt of biopsy-free PGT approach dates from 2013 (Palini et al., 2013). Pailini's group obtained DNA from fluid aspirated from the blastocele cavity. They aimed to analyse particular genes to avoid the transmission of single-gene disorders. That initiative did not become routinely established and, nowadays the embryo biopsy-based protocols are the first-election method in the PGT-M framework (De Rycke et al., 2017). Thus, the efforts on developing new emerging non-invasive protocols are focused on chromosome analysis.
The correlation between numerical chromosome abnormalities and early pregnancy loss, in the field of assisted reproduction was initially suggested in the mid-eighties (Seppälä, 1985). In that time, the capital contribution of chromosome aberrations to the preimplantation loss of embryos produced in vitro was evidenced (Plachot et al., 1988; Wimmers and Van der Merwe, 1988) and it was the seed of a new concept that has arrived at the present time as PGT-A. In essence, the idea was that selecting an euploid embryo to transfer could improve clinical outcomes of the assisted reproduction technologies (ART) by avoiding failed pregnancy attempts, miscarriages, and the need of termination of pregnancy in cases of affected foetus. It was logical and extremely attractive concept that caught the attention of the IVF community. In the recent past, the implementation of embryo micromanipulation procedures together with the fast evolution of genomic technologies allowed to pass from the first in situ hybridization (FISH)-based protocols designed to study a few chromosomes, to modern high-throughput massive genome sequencing platforms (globally named “NGS”: next generation sequencing), in just a decade.
Currently, IVF centres worldwide are performing PGT-A on trophectoderm samples, obtained by embryo biopsy, at the blastocyst stage. The randomized control trials (Yang et al., 2012; Scott et al., 2013a,b; Forman et al., 2013; Rubio et al., 2017; Verpoest et al., 2018; Munné et al., 2019), as well as the meta-analysis (Chen et al., 2015; Dahdouh et al., 2015) shown that PGT-A increases implantation rates, ongoing pregnancy rates and live birth rates per embryo transfer in selected (good prognosis) patients. In addition, euploid single embryo transfer significatively reduces multiple pregnancy rates, miscarriage rates and, practically eliminates aneuploid conceptions. Moreover, it decreases the required number of embryo transfers to achieve a pregnancy. Furthermore, chromosome screening of trophectoderm offers the ultimate biomarker of embryo developmental potential better than morphological or morphokinetic criteria. Finally, is important to note that PGT-A helps to reduce drastically the cryopreservation of spare supernumerary viable blastocyst, as only the euploid ones are vitrified for further reproductive purposes, reducing the surplus cryobanked embryos.
However, some authors are critical about PGT-A and claim for additional validation and clinical experimentation (reviewed in: Rosenwaks et al., 2018; Mochizuki and Gleicher, 2020). The PGT-A detractors argue the mitotic errors (whole-chromosome and segmental aneuploidy) and mosaicism could be confined to the trophectoderm, increasing the false positive results. Furthermore, the hypothesis of self-correction of human embryos could explain the high rates of aneuploidy detected in this tissue. In addition, they doubt about the safety of the trophectoderm biopsy and the representativeness of a few cells respecting to the whole embryo. Besides, an accurate technical validation to demonstrate the clinical validity of new methodologies by comparing with blinded positive controls, such as cell lines, was also claimed. Finally, the irruption of new NGS platforms has evidenced conditions as mosaicism and segmental aneuploidy which have forced to redefine the threshold of “chromosome normality” of human preimplantation embryos.
In the middle of this debate, new forms of non-invasive sampling are being evaluated. The aim of this work is to review the results of the validation and proofs-of-concept experiments reported to date, to stablish the current status of this techniques prior to translation to clinical practice.
The non-invasive PGT approachesThe interest on investigating alternatives to embryo biopsy seems to be very reasonable to extent the applicability of PGT by reducing investment in equipment, time for sample collection and avoiding affecting embryo viability. Additionally, non-invasive methods could reduce both the cost-efficiency of the PGT procedure and human-factor dependency as no experienced embryologists with special skills would be required. In this way, two ni-PGT methods have been proposed to infer the genetic and chromosome status of the embryo.
DNA from blastocoelic fluidThis is not a new idea. In fact, cell-free DNA (cf-DNA) was obtained from liquid aspirated from the blastocoel cavity from day-5 expanded blastocysts in 2013 (Palini et al., 2013). The pioneers of this method proposed the term “blastocentesis” to refer to the aspiration of blastocoele fluid with a conventional ICSI (intracytoplasmic sperm injection) micropipette introduced into the blastocoel cavity (Gianaroli et al., 2014). This system was not strictly “non-invasive” and therefore, it was introduced as the first “minimally-invasive” attempting to obtain DNA from expanded blastocysts. In that occasion, picograms of DNA were obtained and real-time PCR was used for molecular amplification of the multicopy TSPYI gene (on the Y chromosome). Thus, gender identification of male embryos concluded that genotyping of specific loci was reliable (Palini et al., 2013). Nonetheless, obtained DNA was degraded and present at not enough quantity for single copy gene analysis. Despite these limitations, preliminary results encouraged other groups, who applied whole genome amplification (WGA) protocols to increase the total DNA quantity (Galluzzi et al., 2015; Zhang et al., 2016; Shangguan et al., 2017). The WGA step is applied when limited quantity of DNA is available in the primary sample (for example, it usually occurs in forensic samples) which acts as a template for replication in an exponential reaction (Kroneis, 2015; Huang et al., 2015; Li et al., 2018; Lin et al., 2017).
The analysis of individual loci from WGA products obtained from blastocoel fluid (BF) shown extremely varying efficiencies between studies. Amplification rates fluctuated between 34.8% and 87.5% (reviewed by Leaver and Wells, 2020). Additionally, ADO (allele drop-out) rates increase in degraded DNA, rising the risk of misdiagnosis or undiagnosed embryos (Piyamongkol et al., 2003). On the other hand, the grade of concordance between BF and the rest of the blastocyst tissues was not stablished in all cases. Conclusively, it is assumed that BF for accurate PGT-M is pending to further validation and several technique aspects should be improved prior to clinical application (Capalbo et al., 2018).
In the field of PGT-A, the use of BF brings conflicting conclusions (reviewed by Farra et al., 2018; Leaver and Wells, 2020). Globally, the main limitation is the lack of reproducibility among laboratories. There were laboratories reporting WGA amplification and aneuploid/euploid concordance rates (among BF and polar bodies, blastomeres and trophectoderm) up to 90% considering the results as “whole aneuploidy” (Gianaroli et al., 2014; Magli et al., 2016, 2018). However, not full chromosome formula's matching between BF and embryo biopsied samples were reported in all the cases (Gianaroli et al., 2014). Additionally, the analysis of BF was performed by combining WGA protocols with comparative genome hybridization microarray (aCGH). This technique detects copy number variations on whole chromosome but does not accurately detect segmental aneuploidy or mosaicism. Therefore, additional studies established on high-resolution based technologies (as NGS) could probably help to increase the evidence to validate BF results as a genetic test to assist on euploid embryo selection. In any case, the use of BF to PGT-A requires additional validation prior to clinical application because of firstly, the aspiration of BF is not trivial, and embryologist must acquire special skills (Poli et al., 2015; Magli et al., 2016); secondly, the DNA quantity and level of degradation in BF samples varies notably and it is detrimental for standardization. Finally, the improvement diagnostic efficiency depends on the application of more powered genomic tools for chromosome study. In conclusion, genetic analysis of DNA from BF is not right-now a clinically validated tool for preimplantation embryo genetic diagnosis or screening.
DNA from culture mediumThe existence of cell-free DNA (cf-DNA) circulating in blood was reported in 1984 (Mandel and Metais, 1948). Since then, huge amount of work has been addressed to design diagnostic strategies based on the genetic/genomic analysis of this cf-DNA. It was particularly relevant in oncology, because it is possible to obtain valuable clinical information without tumor invasion. In this context, the term “liquid biopsy” has been coined in order to express the method to obtain cf-DNA from blood or any other human fluid. The liquid biopsy opened a new area, the non-invasive genetic diagnosis and screening, that revolutionized the biomedical community. This idea has been recreated in the human embryos growing in vitro. Several authors reported the detection of genomic DNA (gDNA) and mitochondrial DNA (mtDNA) in the medium where embryos were in vitro cultured either up to cleavage-stages as well as to the blastocyst stage (Stigliani et al., 2013, 2014; Assou et al., 2014; Galluzzi et al., 2015; Hammond et al., 2017; Yang et al., 2017). Several reports have been recently published related to technical parameters in the form of both proofs-of-concept and validation works, and two deeply systematic reviews have collected the main conclusions from more than 20 papers on the matter (Leaver and Wells, 2020; Brouillet et al., 2020). In general terms, the great variability reported among groups is noteworthy. From our insight, there are three critical points that merit special attention prior to treating the applicability of cf-DNA on the culture medium in the PGT framework: the cf-DNA detection rate, contamination with non-embryonic DNA and genetic concordance between samples from spent culture medium (SCM) and preimplantation embryo tissues.
DNA detection rateThe reported DNA detection rate varies dramatically from 6.7% (Galluzzi et al., 2015) to 100% (Xu et al., 2016), depending on the technique used for cf-DNA amplification (i.e. quantitative PCR directly or preamplification by WGA plus genomic analysis of the WGA products, respectively). In fact, the efficiency of detection/amplification rates depends on two factors: the protocol used for DNA amplification and the development stage of embryos. Firstly, cf-DNA from SCM amplification is successfully achieved by a great variety of procedures from different providers and with different chemical principles (Farra et al., 2018; Leaver and Wells, 2020; Brouillet et al., 2020). In this sense, WGA protocols merge as the best option for DNA detection/amplification ensuring a wider coverage of the genome (Zong et al., 2012) than PCR-based procedures that are directed to a narrower region of the genome (Voet et al., 2013). However, there is not a universal method to amplify DNA from SCM or limited number of cells. The different WGA procedures have their own advantages and disadvantages in terms of genome coverage, representation bias, error rates, yield and robustness (Deleye et al., 2017). In general terms, the use of MALBAC has been suggested as the preferred amplification method for cf-DNA analyses, because of its high coverage of the genome and low allele drop-out ratio (Zong et al., 2012).
On the other hand, to stablish the best timing for SCM aspiration is crucial to maximize the quantity of sampled DNA. Unfortunately, there is not a consensus on this matter. Lane et al. (2017), demonstrated a higher testing accuracy when SCM sampling occurred on day-4 to day-5 compared to day-3 to day-5. This observation may be explained by the increase on the embryonic:maternal DNA ratio, which occurs coincidentally with the exponential increase of embryonic cell number at blastulation. The exchange or refreshing culture media during the course of embryo development was deemed helpful in eliminating cumulus cells which reduces the impact of maternal contamination. In contrast, other groups (Feichtinger et al., 2017; Hammond et al., 2017) proposed that embryo culture until blastulation, in a continuous system improves the cumulative yield of embryo-derived cf-DNA. On the other hand, other factors do not seem to affect the performance of DNA detection/amplification in the SCM, including the grade of embryo fragmentation, additional embryo manipulation (vitrification, assisted hatching or combined BF plus SCM sampling), the presence of trophectoderm cells out of the zona pellucida (hatched blastocyst) or double amplification strategy (performing of two consecutive WGA rounds on the same sample (Brouillet et al., 2020)).
ContaminationThe detection of exogenous cf-DNA in the intact medium (unexposed to embryos) has been reported by several authors. Brouillet et al. (2020) reported this finding in 57.1% reviewed works (eight out of fourteen) and this phenomenon has been also detected in commercial chemicals used for DNA amplification (personal experience, unpublished data). The existence of non-embryonic cf-DNA in culture medium was deeply analyzed by Hammond et al. (2017) who remarked the presence of baseline DNA contamination in the culture medium non exposed to embryos and concluded that DNA arises throughout the culture period or after sample processing. In addition, they discussed about the confounding role of human serum albumin present in the medium tested, which has a major binding affinity for DNA. The authors claimed the need of implementing adequate controls to fully assess the level of baseline DNA contamination in the culture media in an IVF setting.
On the other hand, Feichtinger et al. (2017) confirmed the presence of maternal DNA contamination in SCM. DNA from cumulus cells has been stressed as the main additional source of contaminating DNA (Capalbo et al., 2018; Rubio et al., 2020). This source of contamination was described as “massive” by Vera-Rodriguez et al. (2018) after finding it in 65% of the samples. Such level of DNA contamination was persistent even after implementing strategies to reduce the background noise of paternal DNA contamination with ‘ICSI-for-all’, carefully removal cumulus cells during the denudation, exchanging the culture media during in vitro incubation or extending the time of culture (Lane et al., 2017). Paternal DNA (sperm cells), laboratory personnel or microorganism's contamination of culture plates cannot be also ruled out. In this sense, the control of all eventual sources of contamination is crucial, and molecular typification of extra-embryonic DNA has not been considered in the majority of studies validating PGT using SCM (Leaver and Wells, 2020). Additional pretesting work-up is mandatory to clarify the mixed origin of DNA extracted from medium.
Concordance of results cf-DNA vs embryonic DNAClinical application of cf-DNA is usually validated comparing genetic results obtained from SCM versus those obtained from preimplantation embryos. At this point, it is necessary to establish what level of concordance is acceptable before its translation to the clinical practice. Besides, the correct template or genetic compartment selected for comparison should be agreed: polar bodies, single blastomere, trophectoderm cells, whole embryo, fetus samples (amniocytes) or newborn samples (peripheral blood, buccal swab) (for overview see Vendrell and Escribà, 2019). Additionally, it is mandatory to unify genetic data to compare genetic variants (genotype), haplotype (short tandem repeats [STR] or single nucleotide polymorphism [SNP]-based), chromosome constitution, total DNA quantity, special DNA fractions (single or double-strand), etc. From this point of view, there is great variability and different comparisons have been stablished among groups. Moreover, the concept of “concordance” has been redefined. Diverse expressions have been used to talk about concordance when results do not exactly match (not perfect chromosome matching achieved): “general concordance”, “full concordance”, “euploid versus aneuploid” or “healthy versus unhealthy”. Following these expressions, the concordance rates varied enormously among groups from 15% to 100% (reviewed by Farra et al., 2018; Brouillet et al., 2020; Leaver and Wells, 2020). In this sense, the concept of concordance goes beyond a semantic matter, and global agreement should be reached in function of the ni-PGT expected scope (diagnosis, screening or indirect estimator of the chromosome status).
It is interesting to explain discordances in order to better understand the inferential power of genetic data obtained from SCM. Besides limitations associated to the origin of the cf-DNA in the SCM (different foreign gDNA has been already discussed), several causes of discordance have been proposed: (i) Source of embryonic DNA: the majority of studies published until now (60%, reviewed by Brouillet et al., 2020) used embryo samples for comparison (polar bodies, single blastomeres or trophectoderm cells) adding difficulty to talk about concordance rates when comparing the genetic results of SCM with a part of the embryo (Gleicher and Barad, 2019). In this sense, discordant result may be the consequence of a different chromosome composition of different parts of the embryo (Soler et al., 2021). At clinical level, whole preimplantation embryos, fetal cells or clinical outcomes from live births seem to be the optimal templates to determine ni-PGT reliability. (ii) Technical challenge: the WGA methods used to amplify cf-DNA from SCM are prone-to-error procedures, introducing bias and sample-to-sample variability that change depending on gDNA base composition content, ADO rates, aberrant preferential allele amplification, dimer (or chimeric) DNA-molecules formation or artificially generated copy errors. These technical limitations should be clarified prior to define the concordance rate. (iii) The origin of cf-DNA: the major limitation concerns to elucidate whether cf-DNA represents the whole embryo genome. Apoptotic or actively dividing embryonic cells release DNA out of the cytoplasm of cells during embryo development. However, inner cell mass and trophectoderm have different division and programed cell-death subroutines. In this case, the exact origin of the cf-DNA found in SCM needs to be still verified. The genetic status of SCM could be inconsistent with the developing preimplantation embryos, therefore it should be taken into account to confirm eventual misdiagnosis due to false negative/positive results.
Applicability of spent embryo culture testingConsidering above cited limitations, the potential use of genetic analysis of cf-DNA from SCM has been tested in both PGT-M and PGT-A frameworks.
PGT-M using spent culture mediumThe use the SCM to detect parental mutations, haplotyping or sex determination of embryos has been reported by some authors (systematically revised by Brouillet et al., 2020) and, once again, there is a great variability concerning embryo manipulation, timing of aspiration, incubation times, amplification methodologies, genetic DNA analysis and concordance. The sex determination was one of the first attempts to test ni-PGT-M after analysis of TSPY1 gene (Y-chromosome gene) (Assou et al., 2014; Galluzzi et al., 2015). In two subsets of analysis the amplification rates varied from 6.7% (2/32) to 38.9%. (21/54) (Galluzzi et al., 2015). Additionally, the amplification product reported of fresh medium (unexposed to embryos) (Hammond et al., 2017) suggesting a DNA contamination within culture media after manufacturing. These data rule out the use of SCM for sex determination and claim the necessity of adequate quality controls to detect false-positive amplification. Any other report has been published to date in relation to the use of SCM for this purpose.
The embryo genotyping, beyond sex determination, has been proposed. In particular, PCR-based methods have been described to determine the carrier status of a deletion variant causing alfa-thalassemia (HBA gene; Wu et al., 2015). The authors obtained very different rates of cf-DNA detection depending on embryo stage (19.67% in day-4 versus 90.16% and 88.46% in day-5 and day-6, respectively). Unfortunately, the concordance between paired samples were not reported and diagnostic evidence remain to be proved. Similarly, Capalbo and co-workers in a technical work (Capalbo et al., 2018) reported an amplification rate of 89.7%, but only 20.8% of samples were concordant with trophectoderm biopsies. In addition, high rates of artifacts/maternal contamination were communicated. Other groups applied WGA plus PCR protocols. Particularly, the detection of C667T polymorphism of the MTHFR gene was reported with an amplification and genotyping rate of 62.5%. Nevertheless, only eight samples were analysed, and solid conclusions were not possible to reach (Galluzzi et al., 2015). In summary, data reported do not provide sufficiently reliable results to support the clinical use of the SCM for PGT-M.
PGT-A using spent culture mediumThe use of SCM for PGT-A has centred the interest of reproductive genetics’ groups. Recently, several reports aimed to validate the clinical applicability of the ni-PGT-A using different designs, laboratory protocols, sampling procedures, additional embryo manipulation, ploidy estimation and concordance criteria. We consider this variability as something usual at these first stages of validation process of new methodology. However, the great variability on data complicates the extrapolation of critical findings and compromise the reproducibility and standardization. The revision of related literature brings sixteen articles with more or less detailed information concerning validation experiments from 2016 to the present day (compiled partially by Farra et al., 2018; Brouillet et al., 2020; Leaver and Wells, 2020). To perform a comprehensive analysis of variability intergroups, we separated three phases in the reported procedures: pre-analytical, analytical and post-analytical phases.
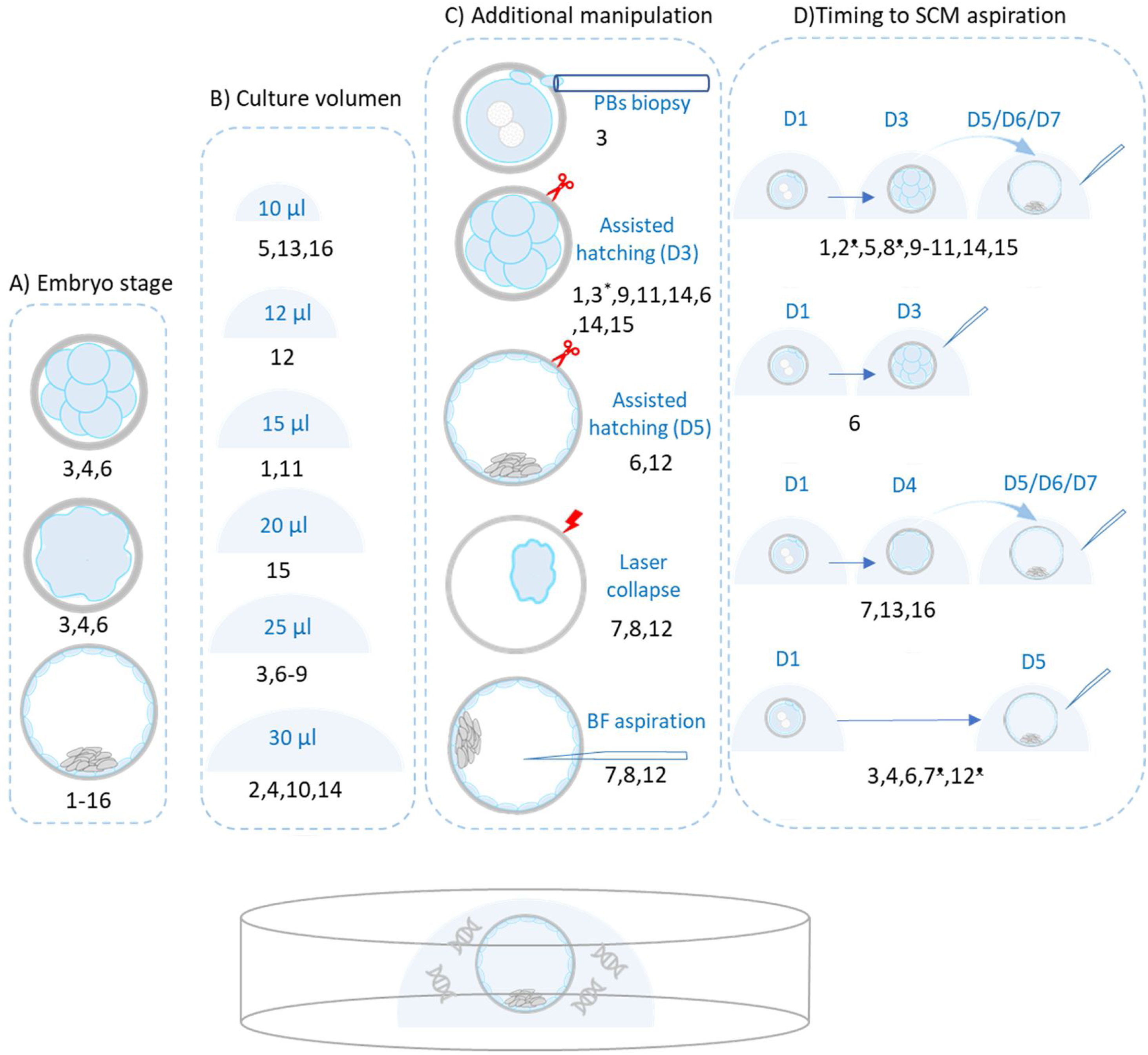
Pre-analytically, we observe a great variability concerning several aspects (Fig. 1): origin of the embryos, fresh/frozen states, additional interventions on embryos, volume of incubation microdrops, type of culture medium and incubation time before SCM collection. Concerning the origin of the embryos, several stages have been used to design comparison experiments: arrested cleavage-stage embryos, arrested blastocyst, expanded spare blastocyst, expanded blastocyst from PGT-A programs or a mix of different stages. Additionally, poor or good-quality embryos are usually mixed. Equally, early, expanded or hatched blastocyst are mixed in some schemes. Thus, diversity of embryo stages implies that comparisons are diverse. Finally, the SCM results are compared with whole-embryos, trophectoderm biopsied cells or disaggregated blastomeres, depending on the experimental design.
The image shows the different experimental designs to validate the chromosome number detection in SCM at the pre-analytical phase. (A) Embryo stage: most groups performed the SCM collection at blastocyst stage and some laboratories aspirated SCM at cleavage or compacting stages. The origin of embryos was diverse, ranging from embryos originated from couples undergoing PGT-A/PGT-M/PGT-SR, supernumerary embryos leftovers from fertility treatments, embryos donated for research following oocyte donation or surplus embryos donated from couples undergoing fertility treatments. In most cases, the final pool of embryos was a mix of fresh and frozen/thawed embryos, at different developmental stages including good and poor-quality embryos, ranging from dividing to early/expanded blastocyst. (B) Culture medium volume ranges from 10 to 30μL. In relation to culture conditions, both sequential and one-step (continuous) culture media from several different commercial compositions are included. (C) Additional embryo manipulation: in spite of the term “non-invasive”, most groups performed some kind of embryo manipulation with the idea of favoring the release of DNA. These manipulations involve biopsy of the polar bodies, assisted hatching on day-3 or day-5 of embryo development, artificial shrinkage of blastocysts by laser pulsing (collapse) or aspiration of fluid from blastocoel cavity to further mixing with SCM collected. (D) Time that embryo spent in culture medium before medium collection. Basically, four strategies have been reported, from the top down: (i) continuous embryo culture to day-3 and subsequent medium refreshing for culture until day 5/6/7, when SCM is collected; (ii) SCM aspiration in day-3; (iii) continuous embryo culture until day-4 and medium refreshing until day 5/6/7, when SCM collection and; (iv) continuous embryo culture until day-5, when SCM is collected. In some cases, the embryos were cryopreserved before the SCM collection. SCM: spent culture medium; BF: blastocoel fluid. *Assisted hatching at 14–18h post-intracytoplasmic sperm injection; •Cryopreserved prior to SCM collection. Numbers correspond to the following references: 1Shamonki et al. (2016); 2Xu et al. (2016); 3Feichtinger et al. (2017); 4Liu et al. (2017); 5Capalbo et al. (2018); 6Ho et al. (2018); 7Kuznyetsov et al. (2018); 8Li et al. (2018); 9Vera-Rodriguez et al. (2018); 10Fang et al. (2019); 11Huang et al. (2019); 12Jiao et al. (2019); 13Rubio et al. (2019); 14Yeung et al. (2019); 15Lledo et al. (2021); 16Rubio et al. (2020).
In relation to fresh or cryopreserved state, both fresh and D2/D3/D5 cryopreserved embryos have been reported. In some case, a mix of both of them (fresh plus frozen/thawed) were included in the same experimental pool (Kuznyetsov et al., 2018). Additionally, in spite of “non-invasive” quality of this strategy, some authors have performed manipulations on the embryos as polar bodies or trophectoderm biopsies, freezing/thawing, assisted hatching, laser collapse or BF aspiration. In some cases, BF and SCM are mixed and jointly analyzed.
In relation to culture volume, it variates from 10μL (Capalbo et al., 2018; Rubio et al., 2019, 2020) to 30μL (Xu et al., 2016; Liu et al., 2017; Fang et al., 2019; Yeung et al., 2019). Different embryo culture schemes from several manufacturers have been registered going from continuous, using one-step medium [G-TL medium (Vitrolife), CSCM (Irvine Scientific), Global (Life Global), Continuous Sage (Origio)] to sequential systems [G1/G2 PLUS (Vitrolife), Quinn (SAGE), Cleavage medium (Cook)+CCM (Vitrolife), Cleavage medium (CooperSurgical)+Quinn (SAGE)]. Concerning the waiting-time before aspiration, in general embryos are washed and changed to fresh-medium drops, where DNA will be finally collected. However, several strategies have been suggested: (i) SCM collection in D3 (Ho et al., 2018); (ii) collection in D5/6/7 (Liu et al., 2017; Feichtinger et al., 2017; Ho et al., 2018; Huang et al., 2019; Jiao et al., 2019). (iii) Changeover step in D3 and collection in D5/D6/D7 (Shamonki et al., 2016; Xu et al., 2016; Li et al., 2018; Vera-Rodriguez et al., 2018; Fang et al., 2019; Yeung et al., 2019; Lledo et al., 2021). (iv) Changeover step in D4 and collection in D5/D6/D7 (Rubio et al., 2019, 2020; Kuznyetsov et al., 2018). Therefore, the secretion time variates from 1 day, 2–3 days, 4 days to 5 days. Finally, most of SCM samples are shipped to external genetic laboratories and scarce information was provided about shipping conditions.
In the analytical phase, aspirated SCM microdrops, are individually processed with commercial WGA kits to amplify the cf-DNA, this technical step is generalized in all reviewed reports. However, modifications on WGA original protocols (Ho et al., 2018; Huang et al., 2019; Jiao et al., 2019; Rubio et al., 2019, 2020) or combination of two different WGA systems in the same experiment have been reported (Vera-Rodriguez et al., 2018). Practically, all commercial kits available have been tested [RepliG (Qiagen), MALBAC and NICS-modified (Yikon Genomics), SurePlex (Illumina), PicoPlex (Takara), NICSwift-modified MALBAC, IonReproseq (ThermoFisher)] to amplify cf-DNA from SCM rendering amplification rates that variated from 82% (Feichtinger et al., 2017) to 100% (Xu et al., 2016; Kuznyetsov et al., 2018; Jiao et al., 2019).
At this point, the quantity of total DNA obtained is variable and depends on the method used to quantify. This issue is relevant because each quantification method has different sensitivity threshold and physicochemical principles [Qubit® (ThermoFisher, fluorometric quantification) 2100 Bioanalyzer® (Agilent, automated electrophoresis); resDNASEQ® (ThermoFisher, quantitative PCR); Nanodrop® (Thermo Scientific, spectrophotometer)]. In this scenario, measured DNA concentration variates considerably from 2 to 642ng/μl (Shamonki et al., 2016). It is challenging to stablish a minimum threshold to distinguish between background noise and really amplified cf-DNA. This is crucial because most of chromosome studies have been performed below the threshold recommended by the manufacturers to perform a copy number chromosome detection. The commercial kits have been designed to trophectoderm biopsies and the minimum quantity of cf-DNA from SCM to initiate the assay is still to be clarified.
In relation to the cytogenetic method used for chromosome analysis, employed platforms go from microarray-based log-ratio estimation [Genetisure (Agilent Technologies) and 24Sure (Illumina)] (Shamonki et al., 2016; Feichtinger et al., 2017) to NGS-based copy number counts per chromosome [NEBnext® and VeriSeq_PGS® (Illumina); Ion Reproseq® (ThermoFisher)]. Again, different NGS platforms are based on different biochemical principles and they have variable resolution rates and coverage of the genome. Two approaches have been used: synthesis-based NGS (Illumina) and ion detection (ThermoFisher). The most widely used method is the high-resolution synthesis-based NGS (Xu et al., 2016; Liu et al., 2017; Li et al., 2018; Kuznyetsov et al., 2018; Fang et al., 2019; Huang et al., 2019; Jiao et al., 2019; Yeung et al., 2019; Lledo et al., 2021) versus ion detection (Ho et al., 2018; Vera-Rodriguez et al., 2018; Rubio et al., 2019, 2020).
Post-analytically, concordance between SCM results and preimplantation embryos changes among revised works. Globally, two forms of reporting concordance have been used: “overall ploidy”, considering the chromosome results as a whole (“normal” vs “abnormal”, “euploid” vs “aneuploid”, “aneuploid” vs “aneuploid” and “euploid vs euploid”) and “full karyotype matching”, comparing the specific chromosomes implied in aneuploidies. In both scenarios, concordance is reported with or without detailed information, respecting mosaicism and segmental aneuploidy, when NGS was applied. There is not clear information respecting the detection rate of this chromosome events in the papers. On the other hand, as it has already suggested, several comparisons have been referred: SCM vs. polar bodies; SCM vs. disaggregated blastomeres from arrested embryos; SCM vs. trophectoderm samples (one or two) used to conventional PGT-A; SCM vs. whole embryo (arrested during cleavage-stage or expanded blastocyst) or SCM vs. live births. In these circumstances, the establishment of concordance is challenging. The overall concordance fluctuates from minimal values of 15.4% (overall aneuploidy comparing SCM versus whole-embryos) in arrested embryos analyzed by ion detection-NGS (Ho et al., 2018) to 100% (overall aneuploidy comparing SCM with trophectoderm biopsies, four to six cells) in expanded blastocyst analyzed by synthesis-based NGS (Kuznyetsov et al., 2018). One paper, registered full concordance in three cases of live births (Xu et al., 2016), but it only confirms euploidy status. The value of concordance concerning full chromosome matchings oscillates from 5%, after comparing whole-chromosome aneuploidies between SCM and trophectoderm samples analyzed by ion detection-NGS (Vera-Rodriguez et al., 2018), to the maximum registered value of 79% after comparing SCM with whole embryos (including mosaicisms and segmental aneuploidy), analyzed by synthesis-based NGS (Kuznyetsov et al., 2018). In general, the full karyotype matching falls drastically compared to the overall aneuploidy concordance.
According to the extreme variability among the different studies, it is difficult to draw global conclusions about ni-PGT-A. Furthermore, the majority of studies are based in a few number of embryos. Most of the studies reported very low concordances to be accepted in a clinical context. The existence of poorly understood biologic mechanisms as mosaicisms, segmental aneuploidy, self-correction hypothesis or differentiate chromosome distribution among blastocyst compartments could explain these discrepancies. In addition, technical aspects are relevant. The variable performance of WGA methods and external or internal (predominantly maternal) DNA contamination cannot be ignored.
On the other side, the implementation of the ni-PGT involve modifications on the current standard operational processes of embryology laboratories. The effect of reducing the culture volume (until 10μl, in some cases) on the physiology of embryos is pending to study. Aspects as the osmolality are crucial and evidence on how hypertonic media may impair embryo development and negatively affect clinical outcomes have been reported (Mestres et al., 2021). This is particularly relevant in dry-culture systems. Similarly, culture extension beyond day 5 in extremely low volumes is not harmless and medium refreshing is mandatory (Insogna et al., 2021) Otherwise, other authors consider that the extended culture beyond day 6 could even be beneficial for selected patients (Hammond et al., 2017). In this way, eventual adaptation of time-lapse, dry or multi-drop culture systems should be considered. Finally, prior to the clinical implementation of these procedures, it would be highly recommended to set up the minimal threshold of both cf-DNA and WGA products, in order to optimize molecular techniques and establish minimal quality standards to obtain reliable results. On this issue, the predictive positive/negative values and likelihood value remain still to be agreed. These values have a real impact on the clinical practice more than statistical significance determination on the concordance rates between samples.
Where does DNA in culture medium come from?The fact that cf-DNA is present in the SCM is undoubted. However, some questions concerning the origin of this DNA are pending to be solved and probably it should be the main initial questions: which embryonic cells are secreting the cf-DNA to the SCM? and what is known about their characteristics or biological significance?
The elucidation of the origin of cf-DNA has focused the interest of geneticist since 1950s (Vlassov et al., 2007). Several biological pathways actively expel DNA to the extracellular environment and some authors have reviewed source and function of this DNA in different clinical scenarios (Bronkhorst et al., 2016; Aucamp et al., 2018; Szilágyi et al., 2020). We may summarize two main origins: cellular breakdown and active DNA release mechanisms (including vesicular transport of nucleic acids). The difference between each mechanism is crucial talking about live or death cells. In the case of “cellular breakdown mechanisms” we are talking about damaged, dead and/or dying cells and processes as necrosis, apoptosis, chromotripsis, pyroptosis, autophagy and mitotic catastrophe would be implicated sharing common pathways as phagocytosis and lysosomal degradation (Aucamp et al., 2018). On the contrary, active DNA release implies living cells and newly synthesised and proactive functional DNA release. Gahan and Stroun (2010) reported that DNA released from cells can be complexed with glycolipoproteins and associating RNA forming functional complexes (virtosomes) detected in culture medium of healthy cells. The DNA fraction of these complexes is newly synthesised in the nuclei of dividing cells and dead or dying cells do not release these complexes. This DNA has functions as intercellular messenger, homeostatic synchronising cell differentiation or activation/inactivation oncogenic processes. Moreover, a source of extracellular DNA is located inside the extracellular vesicles (EV). All human cells excrete EV-containing DNA during many processes, including cancer, cellular differentiation, stress, activation, senescence, stimulation with cytokines, stimulation by shear force, exposure to ATP, apoptotic cell death, changes in the microenvironment, hypoxia, and malignant transformation. EVs are generally categorised as exosomes (30–100nm), apoptotic bodies (<4000nm), prostasomes (40–490nm), large oncosomes (500–10000nm) and microvesicles (100 and 1000nm), which are sometimes referred to as microparticles or shedding vesicles (Raposo and Stoorvogel, 2013). These functional vesicles are particularly secreted at the blastocyst stage and it is hypothesized that are crucial to embryo–endometrial communication in vivo (Bridi et al., 2020).
Taking all together into account, it seems logical to think that in the SCM there exist extracellular DNA from different sources: live cells, death cells and EVs that would be mixed with contaminants (mother/father, handling or manufacturing process). Nowadays, there is not data concerning characterization of each fraction of cf-DNA. It is challenging to quantify the different sub-sets of cf-DNA and determine whether a particular kind of cf-DNA is preferentially amplified by WGA protocols. In this sense, the inference in terms of healthy/unhealthy or euploid/aneuploid/mosaic embryo from a sample of SCM-DNA could be compromised.
On the other hand, beyond the origin of cf-DNA, it is relevant to study the causes of DNA secretion/extrusion to distinguish between normal (physiological) or pathological DNA release. Some authors have speculated about the apoptosis as the main source of cf-DNA released from blastocyst. Nevertheless, programmed cell death subroutines in human blastocyst is not well-understood. A few works published by Hardy and co-workers in 1980–90s (Hardy, 1997, 1999; Hardy et al., 1989) indicated that apoptosis levels are similar in both inner cell mass (ICM) and trophectoderm (TE) compartments. Taking into account the different size (cellularity) of both tissues (ICM represents approximately 15% of the whole blastocyst size), it is conceivable that in the D5-D6 blastocyst, the majority of apoptotically originated cf-DNA might mainly derive from the trophectoderm lineage. Additionally, evidences of euploid livebirths after transfer of mosaic embryos suggest that “euploidization” mechanism may act in human blastocyst. In mice, the self-correction hypothesis defends that chromosomely abnormal cells, located in the foetal lineage (ICM) are eliminated by apoptosis, while those located in the placental (TE) lineage showing proliferative defects though survive (Orvieto et al., 2020). Furthermore, following animal models (Lin et al., 2017), the delay in blastocyst culture could increase the incidence of apoptosis, decrease the ICM:TE cell ratio and influencing the gene expression and diameter of blastocyst derived from in vitro-produced embryos. These arguments would lead us to believe that DNA from SCM is enriched on apoptotic (fragmented and poor-quality DNA, probably from aneuploid cells), increasing the risk of false positive results in ni-PGT. It is important to note that this fact has not been proven in human embryos.
Other cause of DNA release could be the macroautophagy, the sequestering of portions of cytoplasm into a double-membraned vesicle that fuses with lysosomes to promote degradation of their content. Recently, a very impacting publication has proven the linkage between aneuploidy, autophagy and apoptosis in the preimplantation embryos (Singla et al., 2020). Particularly, it has been established that this mechanism acts correcting aneuploidy in murine blastocyst. It is a process mainly reserved for the quality control and turnover of cytosol or cytoplasmic organelles in mammalian cells. Autophagic removal of nuclei can be lethal, and it has been suggested as a mechanism to correct aneuploidy resulting in the release of DNA to extracellular environment. Autophagy is also specially referred to mitochondria turnover (mitophagy) which result in the release of mtDNA into the plasma (Oka et al., 2012; Rello-Varona et al., 2012). At the human blastocyst, mitochondrion clearance was observed in the trophoblast cells because of lysosomal digestion (Escribá et al., 2006). Previously, it has been interpreted as degradation bodies (Cocero et al., 2002; Sathananthan, 2003; Escribá et al., 2006) whose origin is uncertain, but it has been suggested to be related to apoptotic, abnormal cells and cytoplasmic fragments from the embryo (Levy et al., 2001; Sathananthan et al., 2003) participating in the ubiquitin-mediated degradation of paternal mitochondria carried by the spermatozoa during fertilization (Sutovsky, 2003; Sutovsky et al., 2000, 2001). Summarizing, most probably autophagic mechanisms are responsible of expelling fragmented and poor-quality DNA, probably from aneuploid nuclear embryonic DNA and mtDNA.
The oxidative stress is other of the mechanisms triggering apoptosis and it has been classically studied as a plausible cause of cf-DNA release in the form of both normal and oxidised nuclear and apoptotic DNA fragments (Aucamp et al., 2018). The effect of reactive oxygen species in the human oocyte and embryo was suggested in the nineties as aetiological origin of aneuploidy (Tarín, 1995). Furthermore, the detrimental effect of the oxidative stress has been proven on bovine embryos cultured in vitro (Dallemagne et al., 2018). The oxidative stress as a mechanism inducing apoptosis and other cell cycle disturbances, is yet to be deeply analysed in human embryos developed in vitro. Some authors suggested the concomitant action of oxidative stress with other mechanism in the background of this cf-DNA liberation in stressed conditions. Mitotic catastrophe and chromosome fragmentation are two of these mechanisms, described in cultured cells (Stevens et al., 2011). Both mechanisms take place in the G1 phase, following abnormal division. Abnormal chromosome segregation triggers the DNA degradation. This event is associated with classical apoptosis, as the caspase cascade is activated following entry of multinucleated cells into the G1. Fragmented chromosomes are encapsuled in micronuclei in temporally viable cells that posteriorly die and serve as a source of cf-DNA due to the autophagic process. Cancelling or shortening of G1 and/or G2 checkpoints is an essential step in this phenomenon (Roninson et al., 2001). Coincidently, the lack of cell cycle checkpoints is one of the main definitory traits of early human preimplantation embryos and it has been directly related with mosaicism (Harrison et al., 2000). Additionally, chromotripsis, other poorly understood mechanisms, could explain the chromosome instability and segmental aneuploidy find in human blastocyst, as recently suggested (Escribà et al., 2019).
Finally, normal house-keeping functions as DNA replication, repair, recombination, or transcription can contribute to generate cf-DNA that is extracellularly released. However, there are not available data to estimate the impact of this cf-DNA in SCM. Additional causes of cf-DNA concerns genetic processes as re-replication, endoreplication, gene amplification and transposons (Aucamp et al., 2018). It is relevant to note the role of endoreplication in human preimplantation embryos. Endoreplication consists of chromosome replication without cell division. Thus, discrete periods of the S and G phase result in cells with a single polyploid nucleus. This mechanism has been related to morulae's formation and cause of aneuploidy in the human embryos (Daughtry and Chavez, 2016). Programmed endoreplication is related to differentiation and the restarting of normal mitosis during preimplantation development (Taylor et al., 2014; Liu, 2020). The resume of normal cell division after endoreplication exhibits chromosome instability and activation of error-prone mitotic cycles activating mitotic catastrophe and cf-DNA release (Fox and Duronio, 2013).
In conclusion, few studies have focused on identifying the main origin of cf-DNA in the SCM and the scientific discussion is inferred from other clinical scenarios. In any case, theoretical sources and causes of cf-DNA released from live and dead/dying cells should be taken into account in order to translate results obtained from SCM analysis. The utility of cf-DNA depends on the aim of the study. In this sense, functional and actively released DNA fraction corresponds to a small fraction of total cf-DNA isolated and usually protected to the DNAses injury (complexed with DNA-binding proteins) to accomplish one biological function (Szilágyi et al., 2020). On the contrary, cf-DNA released via cellular breakdown corresponds to majority cf-DNA fraction in cell systems, usually it is considered as a by-product of the biological waste and comprises naked, fragmented and degraded DNA molecules. The decision of which fraction of cf-DNA to examine for clinical research in the PGT framework will depend on the quantification and identification of cf-DNA patterns. This preclinical research should be priority considered to optimize SCM sample processing.
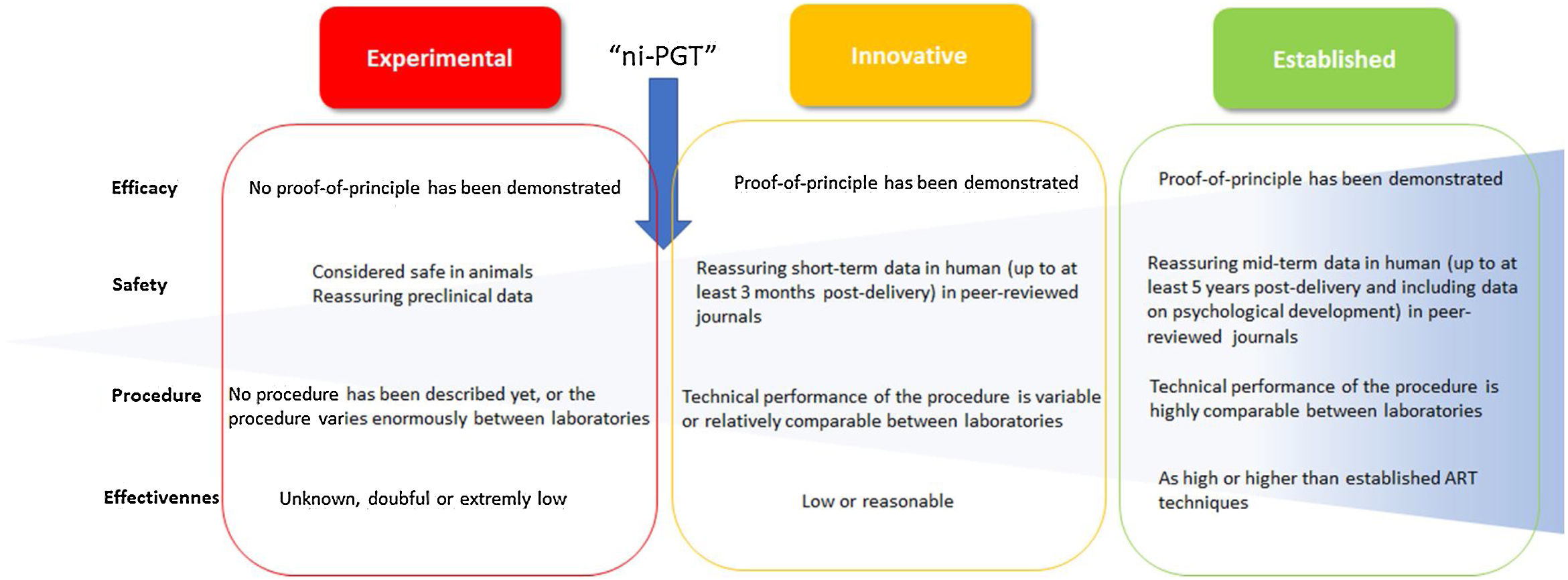
Final remarksThe research of new methods to obtain clinically relevant information from in vitro cultured human embryos has been constant since the beginning of assisted reproduction technologies. Interventions on the embryo or its environment, as well as the interpretation of its morphokinetic behaviour, has offered outstanding data for translational research in our field. In this sense, the experimentation on non-invasive methods to infer the genetic or chromosome status of preimplantation embryos is tremendously attractive. This practice can make PGT accessible to a larger number of IVF laboratories. The genetic analysis of SCM seems to be the most promising approach. Nowadays, it offers solved, unsolved, and seriously limiting issues (Table 1). The efficacy, sensitivity, specificity, reproducibility, and clinical value of the results are yet to be confirmed. Now is the time for these verifications. Reviewing literature, several culture strategies, amplification protocols, and analytical platforms are being tested with the aim to find the best combination to ensure reliable results. The great variability among laboratories is the main issue. From our insight, this variability is assumable at this exploratory stage, however it is necessary to clarify the real state of the technique. Following the boundaries definition between “experimental”, “innovative” and “established” interventions proposed by ESHRE (Provoost et al., 2014) it can be concluded that ni-PGT is at an intermediate stage between experimental and early innovative technique (Fig. 2). In terms of efficacy, the proof-of-principle has been demonstrated and this issue is mandatory to move from experimental to innovative procedures. However, concerning the safety, data from experimental animals and reassuring short-term in human peer-reviewed journals are scarce. Respecting the procedural aspects, the technical performance is highly variable between laboratories, and claims for necessary standardization prior to consider ni-PGT as an innovative intervention. Finally, the effectiveness, defined as the likelihood of producing the desired outcome compared with conventional established ART technique outcome is still low.
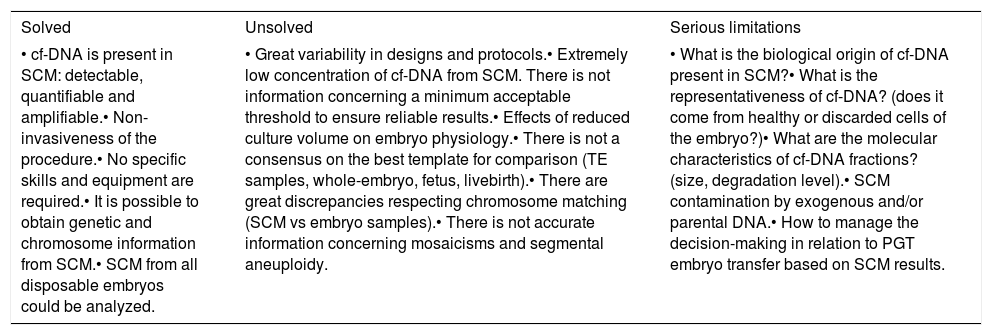
Compilation of solved, unsolved and limiting aspects of the validation of non-invasive preimplantation genetic testing based on spent culture medium. cf-DNA: cell-free DNA; SCM: spent culture medium; TE: trophectoderm.
| Solved | Unsolved | Serious limitations |
|---|---|---|
| • cf-DNA is present in SCM: detectable, quantifiable and amplifiable.• Non-invasiveness of the procedure.• No specific skills and equipment are required.• It is possible to obtain genetic and chromosome information from SCM.• SCM from all disposable embryos could be analyzed. | • Great variability in designs and protocols.• Extremely low concentration of cf-DNA from SCM. There is not information concerning a minimum acceptable threshold to ensure reliable results.• Effects of reduced culture volume on embryo physiology.• There is not a consensus on the best template for comparison (TE samples, whole-embryo, fetus, livebirth).• There are great discrepancies respecting chromosome matching (SCM vs embryo samples).• There is not accurate information concerning mosaicisms and segmental aneuploidy. | • What is the biological origin of cf-DNA present in SCM?• What is the representativeness of cf-DNA? (does it come from healthy or discarded cells of the embryo?)• What are the molecular characteristics of cf-DNA fractions? (size, degradation level).• SCM contamination by exogenous and/or parental DNA.• How to manage the decision-making in relation to PGT embryo transfer based on SCM results. |
Sequential scheme to consider the state of an intervention in the field of the assisted reproduction techniques. The flow-chart shows the evolution of an intervention from “experimental” to “established” status and suggested criteria to be included in each category, considering evidence about efficacy, safety, procedure and effectiveness. The arrow indicates suggested state for non-invasive preimplantation genetic testing (PGT). Adapted from Provoost et al. (2014). ART: assisted reproduction technique.
In conclusion, in agreement with previous authors (Brouillet et al., 2020; Leaver and Wells, 2020; Qasemi et al., 2021) more work is necessary to consider ni-PGT as a clinically indicated technique. Nowadays, the number of studies is limited, and considerable differences exist in design, methods and studied populations. The inferential power of the results obtained from SCM is pending to be confirmed and the decision concerning transfer of an elective blastocyst based on SCM analysis requires of more powerful validation. Mainly, we strongly thought that more data concerning the biological origin and sources of DNA detected in SCM are necessary prior to confirm its representative value. Nevertheless, published data are promising and the effort of groups working on this area is really valuable. Most probably, additional pre-test interventions focused on classifying cf-DNA fractions from SCM as well as monitorization and follow-up of first clinical outcomes are mandatory prior to implement non-invasive PGT into clinical routine practice.
Conflicts of interestThe authors declare no conflicts of interest.